
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,081,751 – 3,081,846 |
| Length | 95 |
| Max. P | 0.930770 |

| Location | 3,081,751 – 3,081,846 |
|---|---|
| Length | 95 |
| Sequences | 11 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 63.52 |
| Shannon entropy | 0.72594 |
| G+C content | 0.46302 |
| Mean single sequence MFE | -29.82 |
| Consensus MFE | -8.04 |
| Energy contribution | -7.87 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.36 |
| SVM RNA-class probability | 0.930770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
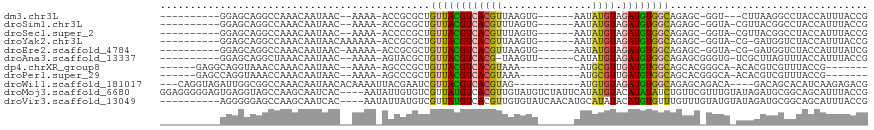
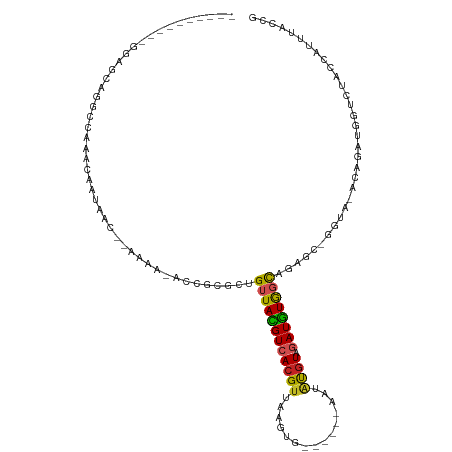
>dm3.chr3L 3081751 95 - 24543557 ----------GGAGCAGGCCAAACAAUAAC--AAAA-ACCGCGCUGUUACGUCACGUUAAGUG------AAUAUGUAGAUGUGGCAGAGC-GGU---CUUAAGGCCUACCAUUUACCG ----------((...(((((..........--....-(((((.((((((((((((((......------...)))).)))))))))).))-)))---.....))))).))........ ( -31.51, z-score = -3.16, R) >droSim1.chr3L 2604597 97 - 22553184 ----------GGAGCAGGCCAAACAAUAAC--AAAA-ACCGCGCUGUUACGUCACGUUUAGUG------AAUAUGUAGAUGUGGCAGAGC-GGUA-CGUUACGGCCUACCAUUUACCG ----------((...(((((......((((--....-(((((.((((((((((((((......------...)))).)))))))))).))-))).-.)))).))))).))........ ( -33.70, z-score = -3.14, R) >droSec1.super_2 3075227 97 - 7591821 ----------GGAGCAGGCCAAACAAUAAC--AAAA-ACCCCGCUGUUACGUCACGUUUAGUG------AAUAUGUAGAUGUGGCAGAGC-GGUA-CGUUACGGCCUACCAUUUACCG ----------((...(((((......((((--....-(((.(.((((((((((((((......------...)))).)))))))))).).-))).-.)))).))))).))........ ( -27.40, z-score = -1.41, R) >droYak2.chr3L 15331782 98 + 24197627 ----------GGAGCAGGCCAAACAAUAACAAAAAA-ACCGCGCUGUUACGUCACGUUAAGUG------AAUAUGUAGAUGUGGCAGAGC-GGUA-CG-GAUGGUCUACCAUUUACCG ----------((........................-(((((.((((((((((((((......------...)))).)))))))))).))-))).-.(-(((((....)))))).)). ( -32.30, z-score = -3.10, R) >droEre2.scaffold_4784 3089916 97 - 25762168 ----------GGAGCAGGCCAAACAAUAAC-AAAAA-ACCGCGCUGUUACGUCACGUUAAGUG------AAUAUGUAGAUGUGGCAGAGC-GGUA-CG-GAUGGUCUACCAUUUAUCG ----------((...((((((..(..(...-...).-(((((.((((((((((((((......------...)))).)))))))))).))-))).-.)-..)))))).))........ ( -31.30, z-score = -2.97, R) >droAna3.scaffold_13337 14257600 97 + 23293914 ----------GGAGCAGGCUAAACAAUAAC--AAAA-AGUACGCUGUUACGUCACG-UAAGUU------CAUAUGUAGAUGUGGCAGAGCGGGUG-UCGCUUAGUUUACCAUUUACCG ----------((....((.(((((......--....-......(((((((((((((-((....------..))))).))))))))))((((....-.))))..))))))).....)). ( -26.30, z-score = -1.38, R) >dp4.chrXR_group8 3944021 91 - 9212921 ------GAGGCAGGUAAACCAAACAAUAAC--AAAA-AGCCCGCUGUUACGUCACGUAAA----------AUGCGUUGAUGUGGCAGCACGGGCA-ACACGUCGUUUACCG------- ------......(((((((...........--....-.....(((((((((((((((...----------..))).))))))))))))((((...-.).))).))))))).------- ( -33.80, z-score = -3.62, R) >droPer1.super_29 175369 91 - 1099123 ------GAGCCAGGUAAACCAAACAAUAAC--AAAA-AGCCCGCUGUUACGUCACGUAAA----------AUGCGUUGAUGUGGCAGCACGGGCA-ACACGUCGUUUACCG------- ------......(((((((...........--....-.....(((((((((((((((...----------..))).))))))))))))((((...-.).))).))))))).------- ( -33.80, z-score = -3.86, R) >droWil1.scaffold_181017 150991 100 - 939937 ---CAGGUAGAUUGGCGGCCAAACAAUAACACAAAAUUACGAAUCGUUACGUCACGUAG-----------AUGUGUAGAUGUGGCAGAGCAGACA----GACAGCACAUCAAGAGACG ---..(((.(.....).))).............................((((.(...(-----------((((((.(.(((.((...))..)))----..).)))))))..).)))) ( -18.10, z-score = 0.08, R) >droMoj3.scaffold_6680 23750356 114 + 24764193 GGAGGGGGAGUGAGGUAGCCAAGCAAUCAC----AAUAUUGUGUCGUUAUGUCACGUUGUAUGUCUAUUCAUAUGUACAUAUAUCUGUUCGUUUGUAUAGAUGCGGCAGCAUUUACCG ...((..((((((.(((((...(((((...----...)))))...))))).))))(((((..((((((.((.(((.(((......))).))).)).))))))...))))).))..)). ( -33.60, z-score = -1.91, R) >droVir3.scaffold_13049 126431 104 - 25233164 ----------AGGGGGAGCCAAGCAAUCAC----AAUAUUAUGUCGUUAUGUCACGUUGUGUAUCAACAUGCAUAUACAUGUGUUUGUUUGUAUGUAUAGAUGCGGCAGCAUUUACCG ----------.((......(((((((.(((----(......(((...((((.((.((((.....)))).)))))).))))))).)))))))......(((((((....))))))))). ( -26.20, z-score = -0.06, R) >consensus __________GGAGCAGGCCAAACAAUAAC__AAAA_ACCGCGCUGUUACGUCACGUUAAGUG______AAUAUGUAGAUGUGGCAGAGC_GGUA_ACAGAUGGUCUACCAUUUACCG .............................................((((((((((((...............)))).))))))))................................. ( -8.04 = -7.87 + -0.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:59:36 2011