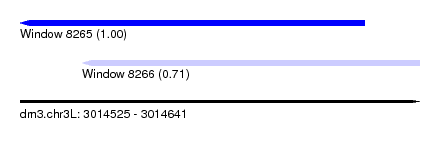
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 3,014,525 – 3,014,641 |
| Length | 116 |
| Max. P | 0.995843 |

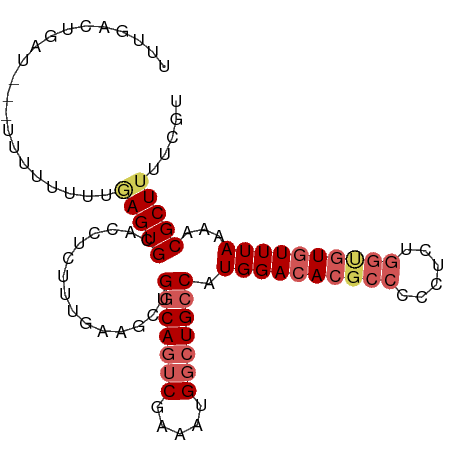
| Location | 3,014,525 – 3,014,625 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 79.64 |
| Shannon entropy | 0.37269 |
| G+C content | 0.45076 |
| Mean single sequence MFE | -28.20 |
| Consensus MFE | -20.68 |
| Energy contribution | -22.24 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.76 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.85 |
| SVM RNA-class probability | 0.995843 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
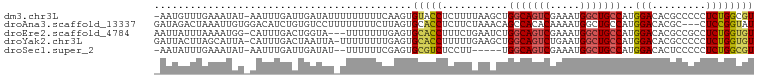
>dm3.chr3L 3014525 100 - 24543557 UUUGAUUGAUAUUUUUUUUUCAAGUGUACCUCUUUUAAGCUGGCAGUCGAAAUGGCUGCCAUGGACACGCCCCCUCUGGCGUGUUUAAAACGCUUUUCGU .((((..((......))..))))((((.............((((((((.....))))))))((((((((((......))))))))))..))))....... ( -32.60, z-score = -3.90, R) >droAna3.scaffold_13337 14197678 95 + 23293914 UCUGUGUCCU--UUUUUUUCUUAGUGCACCUCUUCUAAACAGCCACACAAAAUGGCUGCCAUGGACACG---CCUCCGGUAUCUUUAAAACGCUCUUCGC ...((((((.--........((((..........)))).((((((.......))))))....))))))(---((...))).................... ( -18.90, z-score = -1.58, R) >droEre2.scaffold_4784 3032317 97 - 25762168 UUUGACUGGU---AUUUUUUUGAGUGCACCUUUCUGAAUCUGGCAGUCGAAAUGGCUGCCAUGGACACGCCGCCUCUGGUGUGUUUAAAACGCUUUUCGU ........((---((((....))))))........(((..((((((((.....))))))))(((((((((((....)))))))))))........))).. ( -30.20, z-score = -2.24, R) >droYak2.chr3L 15272683 99 + 24197627 UUUGACUAAUU-AUUUUUUUUGAGUGCACCUUUUUGAAGCUGGCAGUCUGAAUGGCUGCCAUGGACACGCCCCCUCUGGUGUGUUUAAAACGCUUUUCGU ...........-.......................(((((((((((((.....))))))))((((((((((......))))))))))....))))).... ( -30.50, z-score = -3.13, R) >droSec1.super_2 3018670 93 - 7591821 UUUGAUUGAUA--UUUUUUUCGAGUGCGUCUCCUU-----UGGCAGUCGAAAUGGCUGCCAUGGACACUCCCCCUCUGGCGUGUUUAAAACGCUUUUCGU ...........--.......((((.((((......-----((((((((.....))))))))(((((((.((......)).)))))))..))))..)))). ( -28.80, z-score = -2.96, R) >consensus UUUGACUGAU___UUUUUUUUGAGUGCACCUCUUUGAAGCUGGCAGUCGAAAUGGCUGCCAUGGACACGCCCCCUCUGGUGUGUUUAAAACGCUUUUCGU .....................(((((...............(((((((.....))))))).((((((((((......))))))))))...)))))..... (-20.68 = -22.24 + 1.56)

| Location | 3,014,543 – 3,014,641 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 71.59 |
| Shannon entropy | 0.51368 |
| G+C content | 0.42498 |
| Mean single sequence MFE | -23.84 |
| Consensus MFE | -12.46 |
| Energy contribution | -12.78 |
| Covariance contribution | 0.32 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.46 |
| SVM RNA-class probability | 0.705978 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 3014543 98 - 24543557 -AAUGUUUGAAAUAU-AAUUUGAUUGAUAUUUUUUUUUCAAGUGUACCUCUUUUAAGCUGGCAGUCGAAAUGGCUGCCAUGGACACGCCCCCUCUGGCGU -...((((((((...-.((((((..((......))..)))))).......))))))))((((((((.....)))))))).....(((((......))))) ( -27.70, z-score = -2.78, R) >droAna3.scaffold_13337 14197696 97 + 23293914 GAUAGACUAAAUUGUGGACAUCUGUGUCCUUUUUUUUCUUAGUGCACCUCUUCUAAACAGCCACACAAAAUGGCUGCCAUGGACACGC---CUCCGGUAU ..............((((....(((((((.........((((..........)))).((((((.......))))))....))))))).---.)))).... ( -21.20, z-score = -0.79, R) >droEre2.scaffold_4784 3032335 96 - 25762168 AAUUAUUUAAAAUGG-CAUUUGACUGGUA---UUUUUUUGAGUGCACCUUUCUGAAUCUGGCAGUCGAAAUGGCUGCCAUGGACACGCCGCCUCUGGUGU ...........((((-(((((((((((((---(((....)))))).((...........)))))))))).....))))))....((((((....)))))) ( -25.30, z-score = -0.86, R) >droYak2.chr3L 15272701 98 + 24197627 GAUUACUUAGCAUUA-CAUUUGACUAAUUA-UUUUUUUUGAGUGCACCUUUUUGAAGCUGGCAGUCUGAAUGGCUGCCAUGGACACGCCCCCUCUGGUGU .........(((((.-((............-.......))))))).............((((((((.....)))))))).....(((((......))))) ( -22.21, z-score = -0.64, R) >droSec1.super_2 3018688 91 - 7591821 -AAUAUUUGAAAUAU-AAUUUGAUUGAUAU--UUUUUUCGAGUGCGUCUCCUU-----UGGCAGUCGAAAUGGCUGCCAUGGACACUCCCCCUCUGGCGU -..............-.((((((..((...--.))..))))))((((((((..-----((((((((.....)))))))).)))............))))) ( -22.80, z-score = -1.59, R) >consensus _AUUAUUUAAAAUAU_AAUUUGACUGAUAU_UUUUUUUUGAGUGCACCUCUUUGAAGCUGGCAGUCGAAAUGGCUGCCAUGGACACGCCCCCUCUGGUGU ...........................................(((((...........(((((((.....)))))))..(((.........)))))))) (-12.46 = -12.78 + 0.32)
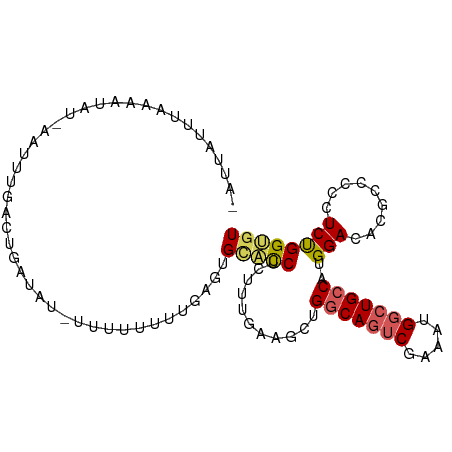
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:59:29 2011