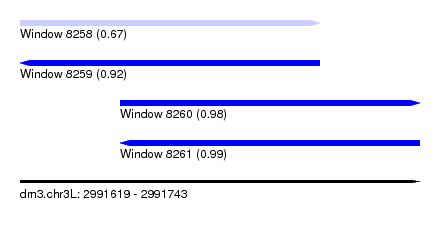
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 2,991,619 – 2,991,743 |
| Length | 124 |
| Max. P | 0.994817 |

| Location | 2,991,619 – 2,991,712 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 93.05 |
| Shannon entropy | 0.11939 |
| G+C content | 0.36533 |
| Mean single sequence MFE | -19.96 |
| Consensus MFE | -18.38 |
| Energy contribution | -17.82 |
| Covariance contribution | -0.56 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.670513 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
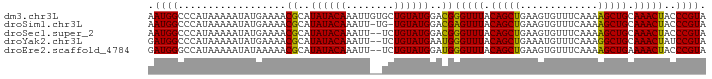
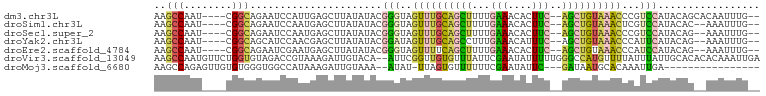
>dm3.chr3L 2991619 93 + 24543557 AAUGGCCCAUAAAAAUAUGAAAACGCAUAUACAAAUUGUGCUGUAUGGACGGGUUUACAGCUGAAGUGUUUCAAAAGCUGCAAACUACCCGUA ......(((((......(....).(((((.......)))))..)))))((((((...(((((.............)))))......)))))). ( -21.72, z-score = -1.08, R) >droSim1.chr3L 2523886 91 + 22553184 AAUGGCCCAUAAAAAUAUGAAAACGCAUAUACAAAUU-UG-UGUAUGGACGAGUUUACAGCUGAAGUGUUUCAAAAGCUGCAAACUACCCGUA .((((..((((....))))......((((((((....-))-))))))....(((((.(((((.............))))).)))))..)))). ( -20.22, z-score = -1.75, R) >droSec1.super_2 2995710 91 + 7591821 AAUGGCCCAUAAAAAUAUGAAAACGCAUAUACAAAUU--UCUGUAUGGACGGGUUUACAGCUGAAGUGUUUCAAAAGCUGCAAACUACCCGUA ......((((....(((((......)))))(((....--..)))))))((((((...(((((.............)))))......)))))). ( -19.92, z-score = -1.26, R) >droYak2.chr3L 15249506 91 - 24197627 GAUGGCCCAUAAAAAUAUGAAAACGCAUAUACAAAUU--UCUGUAUGAAUGGGUUUACAGCUGAAAUGUUUCAAAGGCUGCAAACUAUCCGUA ((((((((((.....((((......))))((((....--..))))...)))))....((((((((....)))...)))))....))))).... ( -17.00, z-score = -0.59, R) >droEre2.scaffold_4784 3009019 91 + 25762168 GAUGGGCCAUAAAAAUAUAAAAACGCAUAUACAAAUU--UCUGUAUGGAUGGGUUUACAGCUGAAGUGUUUCAAAAGCUGAAAACUACCCGUA .(((((.................((..((((((....--..))))))..))(((((.(((((.............))))).))))).))))). ( -20.92, z-score = -2.01, R) >consensus AAUGGCCCAUAAAAAUAUGAAAACGCAUAUACAAAUU__UCUGUAUGGACGGGUUUACAGCUGAAGUGUUUCAAAAGCUGCAAACUACCCGUA .((((..................((..((((((........))))))..))(((((.(((((.............))))).)))))..)))). (-18.38 = -17.82 + -0.56)
| Location | 2,991,619 – 2,991,712 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 93.05 |
| Shannon entropy | 0.11939 |
| G+C content | 0.36533 |
| Mean single sequence MFE | -21.78 |
| Consensus MFE | -18.82 |
| Energy contribution | -18.90 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.922367 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
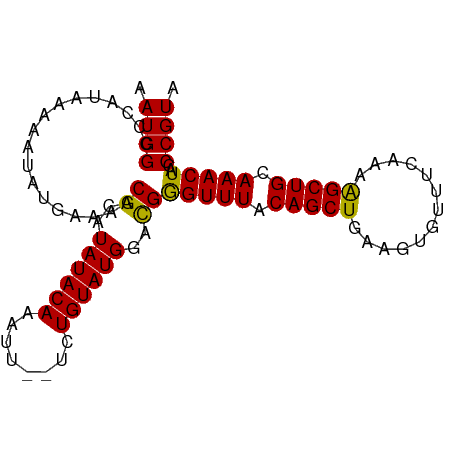
>dm3.chr3L 2991619 93 - 24543557 UACGGGUAGUUUGCAGCUUUUGAAACACUUCAGCUGUAAACCCGUCCAUACAGCACAAUUUGUAUAUGCGUUUUCAUAUUUUUAUGGGCCAUU ...((((...((((((((...(((....)))))))))))))))(((((((..(((.....)))(((((......)))))...))))))).... ( -23.90, z-score = -2.29, R) >droSim1.chr3L 2523886 91 - 22553184 UACGGGUAGUUUGCAGCUUUUGAAACACUUCAGCUGUAAACUCGUCCAUACA-CA-AAUUUGUAUAUGCGUUUUCAUAUUUUUAUGGGCCAUU ....((((((((((((((...(((....))))))))))))))(((..(((((-..-....)))))..)))...(((((....))))))))... ( -23.30, z-score = -2.83, R) >droSec1.super_2 2995710 91 - 7591821 UACGGGUAGUUUGCAGCUUUUGAAACACUUCAGCUGUAAACCCGUCCAUACAGA--AAUUUGUAUAUGCGUUUUCAUAUUUUUAUGGGCCAUU ...((((...((((((((...(((....)))))))))))))))(((((((..((--(((.((............)).)))))))))))).... ( -23.90, z-score = -2.51, R) >droYak2.chr3L 15249506 91 + 24197627 UACGGAUAGUUUGCAGCCUUUGAAACAUUUCAGCUGUAAACCCAUUCAUACAGA--AAUUUGUAUAUGCGUUUUCAUAUUUUUAUGGGCCAUC ...((((.(((((((((...((((....)))))))))))))..))))((((((.--...))))))(((.((..(((((....)))))))))). ( -20.10, z-score = -1.96, R) >droEre2.scaffold_4784 3009019 91 - 25762168 UACGGGUAGUUUUCAGCUUUUGAAACACUUCAGCUGUAAACCCAUCCAUACAGA--AAUUUGUAUAUGCGUUUUUAUAUUUUUAUGGCCCAUC ...((((.((((.(((((...(((....)))))))).))))......((((((.--...)))))).....................))))... ( -17.70, z-score = -1.57, R) >consensus UACGGGUAGUUUGCAGCUUUUGAAACACUUCAGCUGUAAACCCGUCCAUACAGA__AAUUUGUAUAUGCGUUUUCAUAUUUUUAUGGGCCAUU .(((.((((((((((((...((((....))))))))))))).......(((((......)))))))).)))..(((((....)))))...... (-18.82 = -18.90 + 0.08)

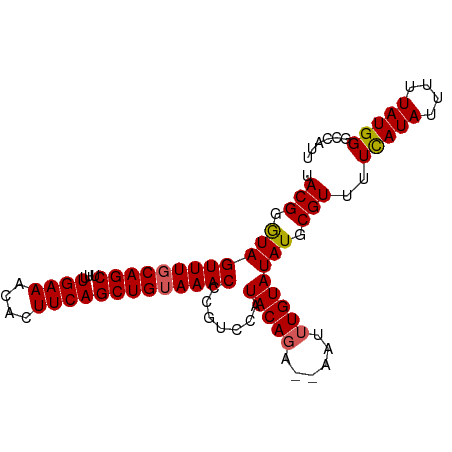
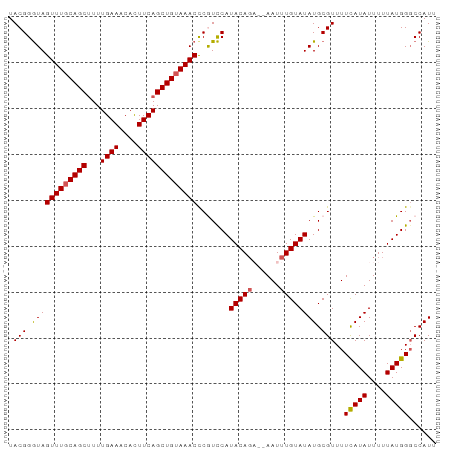
| Location | 2,991,650 – 2,991,743 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 64.83 |
| Shannon entropy | 0.66354 |
| G+C content | 0.38986 |
| Mean single sequence MFE | -21.45 |
| Consensus MFE | -10.04 |
| Energy contribution | -12.07 |
| Covariance contribution | 2.03 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.977301 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
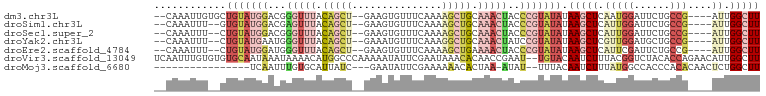
>dm3.chr3L 2991650 93 + 24543557 --CAAAUUGUGCUGUAUGGACGGGUUUACAGCU--GAAGUGUUUCAAAAGCUGCAAACUACCCGUAUAUAAGCUCAAUGGAUUCUGCCG----AUUGGCUU --...((((.(((.((((.((((((...(((((--.............)))))......)))))).))))))).)))).......(((.----...))).. ( -26.42, z-score = -1.73, R) >droSim1.chr3L 2523917 91 + 22553184 --CAAAUUU--GUGUAUGGACGAGUUUACAGCU--GAAGUGUUUCAAAAGCUGCAAACUACCCGUAUAUAAGCUCAUUGGAUUCUGCCG----AUUGGCUU --.....((--((((((((...(((((.(((((--.............))))).)))))..))))))))))(((.(((((......)))----)).))).. ( -29.22, z-score = -3.53, R) >droSec1.super_2 2995741 91 + 7591821 --CAAAUUU--CUGUAUGGACGGGUUUACAGCU--GAAGUGUUUCAAAAGCUGCAAACUACCCGUAUAUAAGCUCAUUGGAUUCUGCCG----AUUGGCUU --.......--.(((((((...(((((.(((((--.............))))).)))))..))))))).(((((.(((((......)))----)).))))) ( -26.92, z-score = -2.51, R) >droYak2.chr3L 15249537 91 - 24197627 --CAAAUUU--CUGUAUGAAUGGGUUUACAGCU--GAAAUGUUUCAAAGGCUGCAAACUAUCCGUAUAUAAGCUCGUUGGAUGCUGCCG----AUUGGCUU --.......--.((((((....(((((.(((((--(((....)))...))))).)))))...)))))).(((((.(((((......)))----)).))))) ( -23.20, z-score = -1.26, R) >droEre2.scaffold_4784 3009050 91 + 25762168 --CAAAUUU--CUGUAUGGAUGGGUUUACAGCU--GAAGUGUUUCAAAAGCUGAAAACUACCCGUAUAUAAGCUCAUUCGAUUCUGCCG----AUUGGCUU --.......--.(((((((...(((((.(((((--.............))))).)))))..))))))).((((.((.(((.......))----).)))))) ( -24.32, z-score = -2.27, R) >droVir3.scaffold_13049 15140220 99 + 25233164 UCAAUUUGUGUGUGCAAUAAAUAAAACAUGGCCCAAAAAUAUUCGAAUAAACACAACCGAAU--UGUACAAUCUUUACGGUCUACACCAGAACAUUGGCUU .(((((((..((((.....................................))))..)))))--))..((((.(((..(((....))).))).)))).... ( -10.69, z-score = 1.39, R) >droMoj3.scaffold_6680 3024406 79 - 24764193 ----------------UCAAUUUGUGCAUUAUC---GAAUAUUCGAAAAAACACUAA-AUAU--UUUACAAUCUUUAUGGCCACCCACACAACUCUGGCUU ----------------...(((((((..((.((---((....)))).))..))).))-))..--..............(((((............))))). ( -9.40, z-score = -1.40, R) >consensus __CAAAUUU__CUGUAUGGACGGGUUUACAGCU__GAAGUGUUUCAAAAGCUGCAAACUACCCGUAUAUAAGCUCAUUGGAUUCUGCCG____AUUGGCUU ............(((((((...(((((.(((((..(((....)))...))))).)))))..))))))).................((((......)))).. (-10.04 = -12.07 + 2.03)
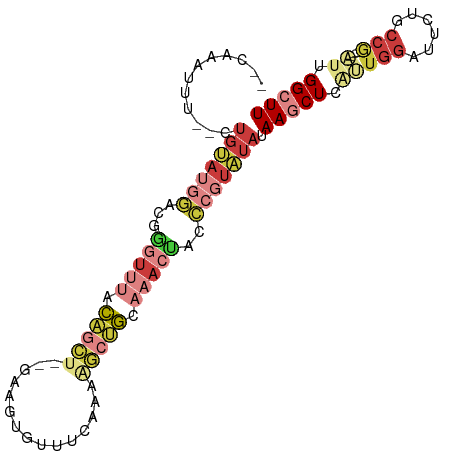
| Location | 2,991,650 – 2,991,743 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 64.83 |
| Shannon entropy | 0.66354 |
| G+C content | 0.38986 |
| Mean single sequence MFE | -20.14 |
| Consensus MFE | -12.07 |
| Energy contribution | -12.00 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.74 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.74 |
| SVM RNA-class probability | 0.994817 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
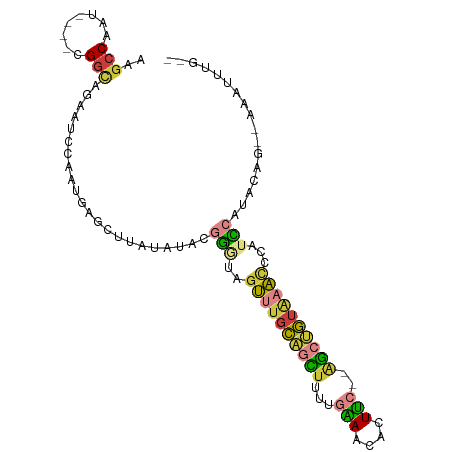
>dm3.chr3L 2991650 93 - 24543557 AAGCCAAU----CGGCAGAAUCCAUUGAGCUUAUAUACGGGUAGUUUGCAGCUUUUGAAACACUUC--AGCUGUAAACCCGUCCAUACAGCACAAUUUG-- ..(((...----.))).......((((.((((((..((((((...((((((((...(((....)))--))))))))))))))..))).))).))))...-- ( -28.40, z-score = -3.88, R) >droSim1.chr3L 2523917 91 - 22553184 AAGCCAAU----CGGCAGAAUCCAAUGAGCUUAUAUACGGGUAGUUUGCAGCUUUUGAAACACUUC--AGCUGUAAACUCGUCCAUACAC--AAAUUUG-- ..(((...----.)))......................(((((((((((((((...(((....)))--))))))))))).))))......--.......-- ( -21.00, z-score = -1.79, R) >droSec1.super_2 2995741 91 - 7591821 AAGCCAAU----CGGCAGAAUCCAAUGAGCUUAUAUACGGGUAGUUUGCAGCUUUUGAAACACUUC--AGCUGUAAACCCGUCCAUACAG--AAAUUUG-- ..(((...----.)))......(((....(((((..((((((...((((((((...(((....)))--))))))))))))))..))).))--....)))-- ( -23.50, z-score = -2.46, R) >droYak2.chr3L 15249537 91 + 24197627 AAGCCAAU----CGGCAGCAUCCAACGAGCUUAUAUACGGAUAGUUUGCAGCCUUUGAAACAUUUC--AGCUGUAAACCCAUUCAUACAG--AAAUUUG-- ..(((...----.)))(((.((....))))).......((((.(((((((((...((((....)))--))))))))))..))))......--.......-- ( -20.80, z-score = -2.49, R) >droEre2.scaffold_4784 3009050 91 - 25762168 AAGCCAAU----CGGCAGAAUCGAAUGAGCUUAUAUACGGGUAGUUUUCAGCUUUUGAAACACUUC--AGCUGUAAACCCAUCCAUACAG--AAAUUUG-- ((((((.(----(((.....)))).)).))))......((((.((((.(((((...(((....)))--))))).))))..))))......--.......-- ( -20.10, z-score = -1.86, R) >droVir3.scaffold_13049 15140220 99 - 25233164 AAGCCAAUGUUCUGGUGUAGACCGUAAAGAUUGUACA--AUUCGGUUGUGUUUAUUCGAAUAUUUUUGGGCCAUGUUUUAUUUAUUGCACACACAAAUUGA ....((((......((((((...((((((...(..((--(((((..((....))..))))).....))..)....))))))...))))))......)))). ( -14.30, z-score = 1.73, R) >droMoj3.scaffold_6680 3024406 79 + 24764193 AAGCCAGAGUUGUGUGGGUGGCCAUAAAGAUUGUAAA--AUAU-UUAGUGUUUUUUCGAAUAUUC---GAUAAUGCACAAAUUGA---------------- .....(.((((.(((((....)))))..)))).)...--....-...((((.((.((((....))---)).)).)))).......---------------- ( -12.90, z-score = -0.01, R) >consensus AAGCCAAU____CGGCAGAAUCCAAUGAGCUUAUAUACGGGUAGUUUGCAGCUUUUGAAACACUUC__AGCUGUAAACCCAUCCAUACAG__AAAUUUG__ ..(((........)))......................(((..((((((((((...(((....)))..))))))))))...)))................. (-12.07 = -12.00 + -0.07)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:59:24 2011