| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,236,372 – 4,236,488 |
| Length | 116 |
| Max. P | 0.644060 |

| Location | 4,236,372 – 4,236,488 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 74.54 |
| Shannon entropy | 0.43953 |
| G+C content | 0.57184 |
| Mean single sequence MFE | -43.63 |
| Consensus MFE | -24.86 |
| Energy contribution | -24.07 |
| Covariance contribution | -0.80 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.644060 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
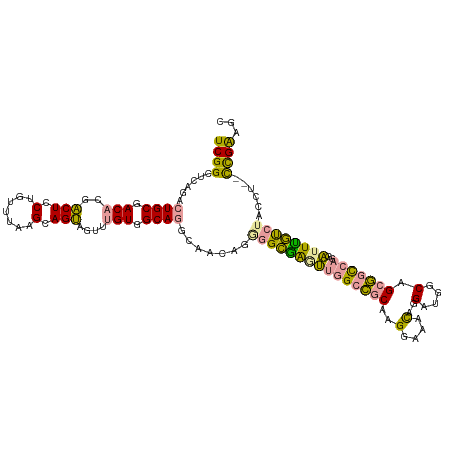
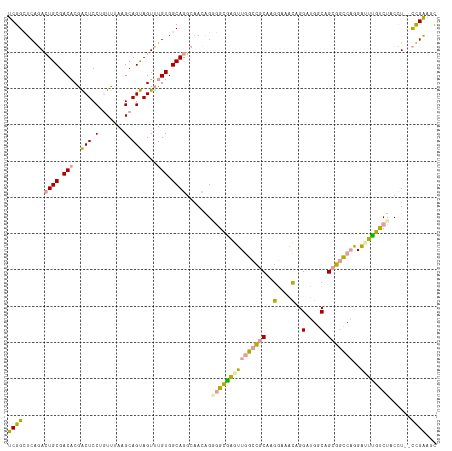
>dm3.chr2L 4236372 116 + 23011544 UCGGCUCAGACUGCGACACGACUCCGGUUUAAGCAGUAGUUUGUGGCAGGCAACAGAGGCGAGUUGGCCGCAAGGAAACAGGAUGGCAGCGGCCAGGAUUUGUCUACCU--CUGAGGC ..(.((((((((((.(((.((((.(.((....)).).))))))).)))).......(((((((((((((((..(....).(.....).)))))))..))))))))...)--))))).) ( -44.30, z-score = -1.68, R) >droSec1.super_5 2333645 116 + 5866729 UCGGCUCAGACUGCGACACGACUCCGGUUUAAGCAGUAGUUUGUGGCAGGCAACAGGGGCGAGUUGGCCGCAAGGAAACAGGAUGGCAGCGGCCAGGAUUUGUCUACCU--CUGGAGC ...((((...((((.(((.((((.(.((....)).).))))))).))))....((((((((((((((((((..(....).(.....).)))))))..)))))....)))--))))))) ( -42.90, z-score = -1.10, R) >droYak2.chr2L 4263349 116 + 22324452 UCGGCUCAGACUGCGACACGACUCCUGUUUAAGCAGUAGUUUGUGGCAGGCAACAGGGGCGAAUUGGCCGCAAGGAAACAGGAUGGCAGCGGCCAGGAUUUGUCUACCU--CCGAAUC ((((......((((.(((.((((.((((....)))).))))))).)))).......(((((((((((((((..(....).(.....).)))))))..))))))))....--))))... ( -44.20, z-score = -2.17, R) >droEre2.scaffold_4929 4309959 116 + 26641161 UCGGCUCAGACUGCGACACGACUCCUGUUUAAGCAGUAGUUUGUGGCAGGCAACAGGGGCAAGUUGGCCGCAAGGAAACAGGAUGGCAGCGGCCAGGAUUUGUCUACCU--CUGAAUC ..(..(((((((((.(((.((((.((((....)))).))))))).)))).......(((((((((((((((..(....).(.....).)))))))..))))))))...)--))))..) ( -45.80, z-score = -2.48, R) >dp4.chr4_group2 226148 116 + 1235136 UCGGAUCGCAGUGCGACUCAGCUCCAAUUUAAGCAGCAGUCUGUGGCAACAGAUUG--GUGGGGUAGCAGCAGGCAGCUGGGGGGACAGGUCUCUCGUGUCACCUUCAUGCUCGAGGU (((....((..(((..((..((((((..........((((((((....))))))))--.))))))))..))).))(((((((((((((.(.....).)))).)))))).))))))... ( -41.20, z-score = -0.38, R) >droPer1.super_8 303573 116 - 3966273 UCGGAUCGCAGUGCGACUCAGCUCCUAUUUAAGCAGCAGUCUGUGGCAACAGAUUG--GUGGGGUAGCAGCAGGCAGCUGGGGGGACAGGUCUCUCGUGUCACCUUCAUGCUCGAGGU .....((((...))))(((.(((.((((((...((.((((((((....))))))))--.)))))))).)))..(.(((((((((((((.(.....).)))).)))))).))))))).. ( -43.40, z-score = -0.83, R) >consensus UCGGCUCAGACUGCGACACGACUCCUGUUUAAGCAGUAGUUUGUGGCAGGCAACAGGGGCGAGUUGGCCGCAAGGAAACAGGAUGGCAGCGGCCAGGAUUUGUCUACCU__CCGAAGC ((((......((((.(((..(((.(.......).)))....))).)))).......(((((((((((((((..(....)..(....).)))))))..))))))))......))))... (-24.86 = -24.07 + -0.80)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:15:45 2011