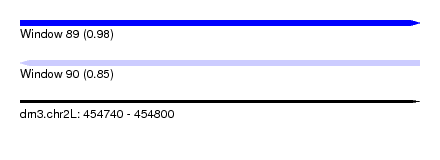
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 454,740 – 454,800 |
| Length | 60 |
| Max. P | 0.975872 |

| Location | 454,740 – 454,800 |
|---|---|
| Length | 60 |
| Sequences | 5 |
| Columns | 60 |
| Reading direction | forward |
| Mean pairwise identity | 68.83 |
| Shannon entropy | 0.56904 |
| G+C content | 0.40158 |
| Mean single sequence MFE | -14.66 |
| Consensus MFE | -7.26 |
| Energy contribution | -8.46 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.975872 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
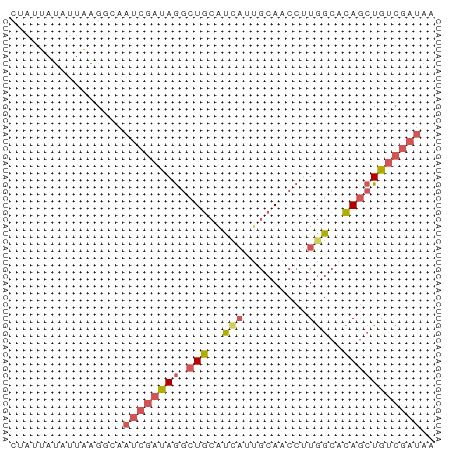
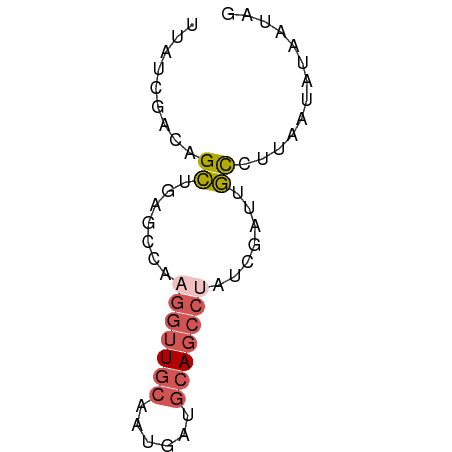
>dm3.chr2L 454740 60 + 23011544 CUAUUAUAUUAAGACAAUCGAUAGGCUGCAUUAUUGCAACCUUGGCUCAGCUGUCGAUAA ................(((((((((.((((....)))).))).(((...))))))))).. ( -13.40, z-score = -1.42, R) >droEre2.scaffold_4929 507295 56 + 26641161 ----UAUAUUAGGGCAAUCGAUAGGCUGCGUCAUUGCAACCGUGGCUCAGCUGUCGAUAA ----............((((((((.(((.(((((.......))))).))))))))))).. ( -18.80, z-score = -2.17, R) >droSec1.super_14 438591 60 + 2068291 CUAUUAUAUUAAGGCAAUCGAUAGGCUGCAUCAUUGCAACCUUGGCACAGCUGUCGAUAA ................((((((((.(((..(((.........)))..))))))))))).. ( -13.80, z-score = -0.98, R) >droSim1.chr2L 461973 60 + 22036055 CUAUUAUAUUAAGGCAAUCGAUAGGCUGCAUCAUUGCAACCUUGGCACAGCUGUCGAUAA ................((((((((.(((..(((.........)))..))))))))))).. ( -13.80, z-score = -0.98, R) >apiMel3.GroupUn 857907 55 + 399230636 AUAGAACAUUUGGAUGGUC-AUAAAGUGACUAUUCAAAGGGAUAAAGUAACUAAAC---- ......(.(((((((((((-((...))))))))))))).)................---- ( -13.50, z-score = -3.95, R) >consensus CUAUUAUAUUAAGGCAAUCGAUAGGCUGCAUCAUUGCAACCUUGGCACAGCUGUCGAUAA ................((((((((.(((..(((.........)))..))))))))))).. ( -7.26 = -8.46 + 1.20)


| Location | 454,740 – 454,800 |
|---|---|
| Length | 60 |
| Sequences | 5 |
| Columns | 60 |
| Reading direction | reverse |
| Mean pairwise identity | 68.83 |
| Shannon entropy | 0.56904 |
| G+C content | 0.40158 |
| Mean single sequence MFE | -12.38 |
| Consensus MFE | -6.08 |
| Energy contribution | -6.92 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.41 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.850685 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 454740 60 - 23011544 UUAUCGACAGCUGAGCCAAGGUUGCAAUAAUGCAGCCUAUCGAUUGUCUUAAUAUAAUAG .....(((((.(((....((((((((....)))))))).))).)))))............ ( -18.10, z-score = -3.54, R) >droEre2.scaffold_4929 507295 56 - 26641161 UUAUCGACAGCUGAGCCACGGUUGCAAUGACGCAGCCUAUCGAUUGCCCUAAUAUA---- ..(((((..((...))...((((((......))))))..)))))............---- ( -12.00, z-score = -0.85, R) >droSec1.super_14 438591 60 - 2068291 UUAUCGACAGCUGUGCCAAGGUUGCAAUGAUGCAGCCUAUCGAUUGCCUUAAUAUAAUAG ..(((((..((...))..((((((((....)))))))).)))))................ ( -13.80, z-score = -1.15, R) >droSim1.chr2L 461973 60 - 22036055 UUAUCGACAGCUGUGCCAAGGUUGCAAUGAUGCAGCCUAUCGAUUGCCUUAAUAUAAUAG ..(((((..((...))..((((((((....)))))))).)))))................ ( -13.80, z-score = -1.15, R) >apiMel3.GroupUn 857907 55 - 399230636 ----GUUUAGUUACUUUAUCCCUUUGAAUAGUCACUUUAU-GACCAUCCAAAUGUUCUAU ----..................((((.((.((((.....)-))).)).))))........ ( -4.20, z-score = -0.35, R) >consensus UUAUCGACAGCUGAGCCAAGGUUGCAAUGAUGCAGCCUAUCGAUUGCCUUAAUAUAAUAG .........((.......(((((((......))))))).......))............. ( -6.08 = -6.92 + 0.84)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:05:54 2011