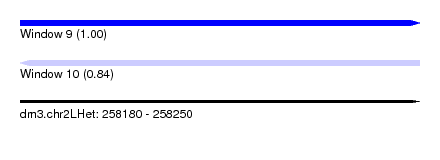
| Sequence ID | dm3.chr2LHet |
|---|---|
| Location | 258,180 – 258,250 |
| Length | 70 |
| Max. P | 0.999720 |

| Location | 258,180 – 258,250 |
|---|---|
| Length | 70 |
| Sequences | 5 |
| Columns | 70 |
| Reading direction | forward |
| Mean pairwise identity | 58.35 |
| Shannon entropy | 0.74354 |
| G+C content | 0.43583 |
| Mean single sequence MFE | -11.24 |
| Consensus MFE | -6.58 |
| Energy contribution | -7.22 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.25 |
| SVM RNA-class probability | 0.999720 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
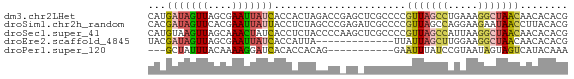
>dm3.chr2LHet 258180 70 + 368872 CAUGAUAGUUAGCGAAUUAUCACCACUAGACCGAGCUCGCCCCGUUAGCCUGAAAGGCUAACAACACACG ..((((((((....)))))))).....................(((((((.....)))))))........ ( -16.40, z-score = -2.86, R) >droSim1.chr2h_random 2530038 70 - 3178526 CACGAUAGUUCACGAAUUAUUACCUCUAGCCCGAGAUCGCCCCGUUAGCCAGGAAGAAUAACCUUACACG ...(((((((....))))))).((((((((.((....))....)))))..)))................. ( -8.50, z-score = -0.28, R) >droSec1.super_41 58351 70 + 296808 CAUGUAAGUUAGCAAACUAUCACCUCUACCCCAAGCUCGCCCCGUUAGCCAUUAAGGCUAACAACACACG ..(((..((.(((.....................))).))...(((((((.....))))))).))).... ( -13.10, z-score = -3.12, R) >droEre2.scaffold_4845 261897 57 + 22589142 UACGAUAGUUAGCGAAUUAUCACCAUUA-------------UUAUUAGCUUGGAAGGCUAACAACACACG ...(((((((....))))))).......-------------...((((((.....))))))......... ( -9.80, z-score = -2.49, R) >droPer1.super_120 50364 56 - 124538 ---GCUAUUUACAAAAGGAUCACACCACAG-----------GAAUUUAUCCGUAAUAGUAGUCAUACAAA ---((((((.((....((......))...(-----------((.....)))))))))))........... ( -8.40, z-score = -2.44, R) >consensus CACGAUAGUUAGCGAAUUAUCACCACUAGCCC_AG_UCGCCCCGUUAGCCAGGAAGGCUAACAACACACG ...(((((((....)))))))......................(((((((.....)))))))........ ( -6.58 = -7.22 + 0.64)

| Location | 258,180 – 258,250 |
|---|---|
| Length | 70 |
| Sequences | 5 |
| Columns | 70 |
| Reading direction | reverse |
| Mean pairwise identity | 58.35 |
| Shannon entropy | 0.74354 |
| G+C content | 0.43583 |
| Mean single sequence MFE | -16.89 |
| Consensus MFE | -5.28 |
| Energy contribution | -6.68 |
| Covariance contribution | 1.40 |
| Combinations/Pair | 1.64 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.87 |
| SVM RNA-class probability | 0.840669 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2LHet 258180 70 - 368872 CGUGUGUUGUUAGCCUUUCAGGCUAACGGGGCGAGCUCGGUCUAGUGGUGAUAAUUCGCUAACUAUCAUG ..(((.(((((((((.....))))))))).))).........(((((((((....))))).))))..... ( -22.90, z-score = -2.19, R) >droSim1.chr2h_random 2530038 70 + 3178526 CGUGUAAGGUUAUUCUUCCUGGCUAACGGGGCGAUCUCGGGCUAGAGGUAAUAAUUCGUGAACUAUCGUG .((....(((((((((((.(((((..((((.....))))))))))))).))))))).....))....... ( -15.50, z-score = 0.15, R) >droSec1.super_41 58351 70 - 296808 CGUGUGUUGUUAGCCUUAAUGGCUAACGGGGCGAGCUUGGGGUAGAGGUGAUAGUUUGCUAACUUACAUG (((((((((((((((.....)))))))))((((((((...............))))))))....)))))) ( -23.66, z-score = -2.74, R) >droEre2.scaffold_4845 261897 57 - 22589142 CGUGUGUUGUUAGCCUUCCAAGCUAAUAA-------------UAAUGGUGAUAAUUCGCUAACUAUCGUA ....((((((((((.......))))))))-------------)).((((((....))))))......... ( -14.50, z-score = -3.12, R) >droPer1.super_120 50364 56 + 124538 UUUGUAUGACUACUAUUACGGAUAAAUUC-----------CUGUGGUGUGAUCCUUUUGUAAAUAGC--- .......((.(((.((((((((.....))-----------).)))))))).))..............--- ( -7.90, z-score = -0.43, R) >consensus CGUGUGUUGUUAGCCUUACAGGCUAACGGGGCGA_CU_GGGCUAGAGGUGAUAAUUCGCUAACUAUCAUG ...((.(((((((((.....))))))))).))...................................... ( -5.28 = -6.68 + 1.40)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:04:48 2011