
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 2,958,548 – 2,958,645 |
| Length | 97 |
| Max. P | 0.967334 |

| Location | 2,958,548 – 2,958,645 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 63.13 |
| Shannon entropy | 0.73143 |
| G+C content | 0.47057 |
| Mean single sequence MFE | -30.12 |
| Consensus MFE | -11.64 |
| Energy contribution | -12.78 |
| Covariance contribution | 1.15 |
| Combinations/Pair | 1.59 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.78 |
| SVM RNA-class probability | 0.967334 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
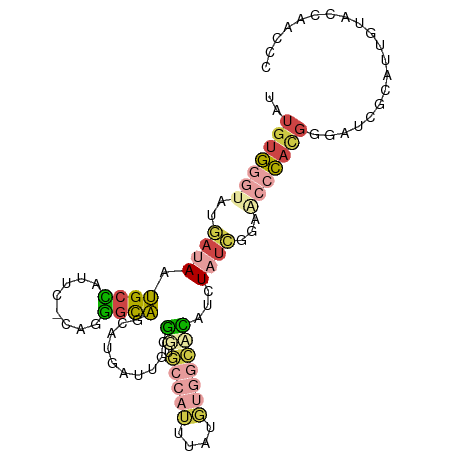
>dm3.chr3L 2958548 97 + 24543557 UAUGUGGGUAUGAUAAUGCCAUUC-UAGGGCAGCAUGAUUGCGGUGCCAUUUAUGUGGCCCAUCUAUCGGAGCCCAAGGGAUCGCAUUGUACCAACCC .....((((...(((((((.((((-(.((((.(((....)))((.(((((....)))))))..........))))..))))).)))))))....)))) ( -37.80, z-score = -2.15, R) >droSim1.chr3L 2498174 97 + 22553184 UAUGUGGGUAUGAUAAUGCCAUUC-GAGGGCAGCCUGAUUGCGGUGCCAUUUAGGUGGCCCAUCUAUCGGAACCCACGGGAUCGCAUUGUACCAACCC .....((((...(((((((.((((-((((((.((((((.((......)).)))))).)))).))...((.......)))))).)))))))....)))) ( -33.40, z-score = -0.67, R) >droSec1.super_2 2969912 97 + 7591821 UAUGUGGGUAUGAUAAUGCCAUUC-GAGGGCAGCCUGCUUGCGGUGCCAUUUAGGUGGCCCAUCUAUCGGAACCCACGGGAUCGCAUUGUAGCAACCC ...((((((.(((((..(((((..-(((((((.((.......)))))).)))..))))).....)))))..))))))(((...((......))..))) ( -34.60, z-score = -0.70, R) >droYak2.chr3L 15220807 77 - 24197627 UAUGUGGGCAACCGAAUGCUUUUC-UAGGGUA-------UGUG-------------AGUAUAUAUAUCGCAACCCACGGAAUCGCAUUCUAACCACCC .....((....))((((((..(((-(.((((.-------((((-------------(.........)))))))))..))))..))))))......... ( -26.10, z-score = -3.37, R) >droEre2.scaffold_4784 2980873 96 + 25762168 UAUGUGGGUGAGAUAAUGCCUUUC-CAGGGUA-UGUGGUUGCGUUGCCAUCUAUGUGGCACAUCUAUCGAAAAACACGGGAUCGCAUUGUACCAACCC .....((((...(((((((..(((-(..((((-.(((((..(((........)))..)).))).))))(.....)..))))..)))))))....)))) ( -28.30, z-score = -0.55, R) >droAna3.scaffold_13337 14152586 96 - 23293914 UAUGUAUCCAGUACAAGGUGUUUCACAACUUUUCAUUCAAUUUCUUUCCAUGAUUUUCGAAGUUUAUAGAUACUUUUCAGAUUUUUGUACUGGAAC-- ......(((((((((((((((((...((((((....(((...........))).....))))))...)))))))..........))))))))))..-- ( -20.50, z-score = -2.62, R) >consensus UAUGUGGGUAUGAUAAUGCCAUUC_CAGGGCAGCAUGAUUGCGGUGCCAUUUAUGUGGCACAUCUAUCGGAACCCACGGGAUCGCAUUGUACCAACCC ..(((((((..((((.((((........))))..........((.(((((....)))))))...))))...))))))).................... (-11.64 = -12.78 + 1.15)

| Location | 2,958,548 – 2,958,645 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 63.13 |
| Shannon entropy | 0.73143 |
| G+C content | 0.47057 |
| Mean single sequence MFE | -29.19 |
| Consensus MFE | -10.10 |
| Energy contribution | -12.07 |
| Covariance contribution | 1.96 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.932804 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
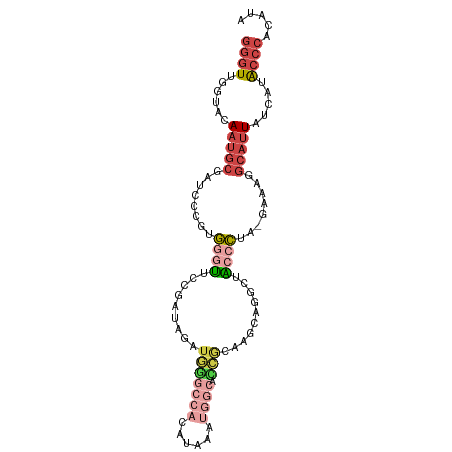
>dm3.chr3L 2958548 97 - 24543557 GGGUUGGUACAAUGCGAUCCCUUGGGCUCCGAUAGAUGGGCCACAUAAAUGGCACCGCAAUCAUGCUGCCCUA-GAAUGGCAUUAUCAUACCCACAUA (((((((((..((((.((..((.((((..........((((((......)))).))(((....))).)))).)-).)).))))))))).))))..... ( -33.60, z-score = -1.54, R) >droSim1.chr3L 2498174 97 - 22553184 GGGUUGGUACAAUGCGAUCCCGUGGGUUCCGAUAGAUGGGCCACCUAAAUGGCACCGCAAUCAGGCUGCCCUC-GAAUGGCAUUAUCAUACCCACAUA (((((((((..((((.((..((.((((.(((((.(.(((((((......)))).)))).))).))..)))).)-).)).))))))))).))))..... ( -36.70, z-score = -1.86, R) >droSec1.super_2 2969912 97 - 7591821 GGGUUGCUACAAUGCGAUCCCGUGGGUUCCGAUAGAUGGGCCACCUAAAUGGCACCGCAAGCAGGCUGCCCUC-GAAUGGCAUUAUCAUACCCACAUA (((((((......))))))).((((((...(((((....((((.......((((((.......)).))))...-...)))).)))))..))))))... ( -36.62, z-score = -1.91, R) >droYak2.chr3L 15220807 77 + 24197627 GGGUGGUUAGAAUGCGAUUCCGUGGGUUGCGAUAUAUAUACU-------------CACA-------UACCCUA-GAAAAGCAUUCGGUUGCCCACAUA (((..(...((((((..(((...((((((.((.........)-------------).))-------.))))..-)))..))))))..)..)))..... ( -25.60, z-score = -2.47, R) >droEre2.scaffold_4784 2980873 96 - 25762168 GGGUUGGUACAAUGCGAUCCCGUGUUUUUCGAUAGAUGUGCCACAUAGAUGGCAACGCAACCACA-UACCCUG-GAAAGGCAUUAUCUCACCCACAUA ((((.((...(((((......(((((..((....))...((((......)))))))))..(((..-.....))-)....)))))..)).))))..... ( -23.30, z-score = -0.04, R) >droAna3.scaffold_13337 14152586 96 + 23293914 --GUUCCAGUACAAAAAUCUGAAAAGUAUCUAUAAACUUCGAAAAUCAUGGAAAGAAAUUGAAUGAAAAGUUGUGAAACACCUUGUACUGGAUACAUA --((((((((((((...............................((((...((....))..))))...(((....)))...)))))))))).))... ( -19.30, z-score = -2.45, R) >consensus GGGUUGGUACAAUGCGAUCCCGUGGGUUCCGAUAGAUGGGCCACAUAAAUGGCACCGCAAGCAGGCUACCCUA_GAAAGGCAUUAUCAUACCCACAUA ((((......(((((........((((.........(((((((......)))).)))..........))))........))))).....))))..... (-10.10 = -12.07 + 1.96)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:59:17 2011