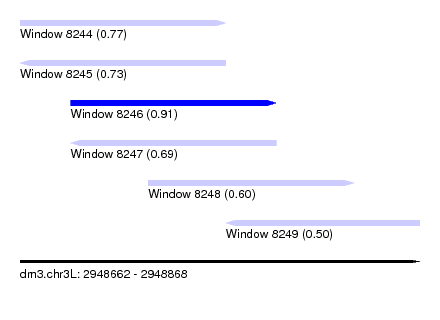
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 2,948,662 – 2,948,868 |
| Length | 206 |
| Max. P | 0.910635 |

| Location | 2,948,662 – 2,948,768 |
|---|---|
| Length | 106 |
| Sequences | 9 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 70.39 |
| Shannon entropy | 0.58609 |
| G+C content | 0.62468 |
| Mean single sequence MFE | -42.43 |
| Consensus MFE | -25.85 |
| Energy contribution | -26.10 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.38 |
| Mean z-score | -0.72 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.772002 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
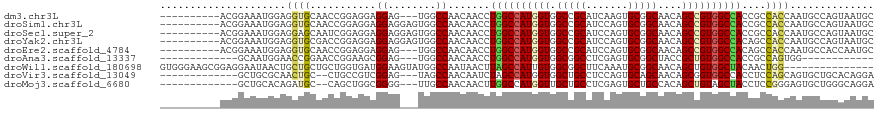
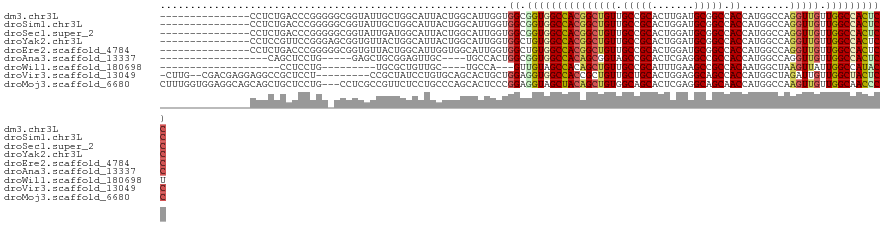
>dm3.chr3L 2948662 106 + 24543557 ----------ACGGAAAUGGAGGUGCAACCGGAGGAGGAG---UGGCCAACAACCUGGCCAUGGUGGCCGCAUCAAGUGCGGCAACAGCCGUGGCCACCGCCACCAAUGCCAGUAAUGC ----------..((...(((.(((....((......)).(---((((((......)))))))(((((((((((...))))(((....)))..)))))))))).)))...))........ ( -46.60, z-score = -1.86, R) >droSim1.chr3L 2488377 109 + 22553184 ----------ACGGAAAUGGAGGUGCAACCGGAGGAGGAGGAGUGGCCAACAACCUGGCCAUGGUGGCCGCAUCCAGUGCGGCAACAGCCGUGGCCACCGCCACCAAUGCCAGUAAUGC ----------..((...(((.(((....((......))....(((((((......)))))))(((((((((((...))))(((....)))..)))))))))).)))...))........ ( -46.60, z-score = -1.30, R) >droSec1.super_2 2960073 109 + 7591821 ----------ACGGAAAUGGAGGAGCAAUCGGAGGAGGAGGAGUGGCCAACAACCUGGCCAUGGUGGCCGCAUCCAGUGCGGCAACAGCCGUGGCCACCGCCACCAAUGCCAGUAAUGC ----------..((...(((..((....))((.((...(((..((.....)).)))(((((((((.(((((((...)))))))....))))))))).)).)).)))...))........ ( -45.80, z-score = -1.69, R) >droYak2.chr3L 15210655 109 - 24197627 ----------ACGGAAAUGGAGGUGCGACCGGAGGAGGAGGAGUGGCCAACAACCUGGCCAUGGUGGCCGCAUCCAGUGCGGCAACAGCCGUGGCCACAGCCACCAAUGCCAGUAAUGC ----------..((...(((.(((....((......))(((..((.....)).)))(((((((((.(((((((...)))))))....)))))))))...))).)))...))........ ( -44.60, z-score = -0.98, R) >droEre2.scaffold_4784 2970894 106 + 25762168 ----------ACGGAAAUGGAGGUGCAACCGGAGGAGGAG---UGGCCAACAACCUGGCCAUGGUGGCCGCAUCCAGUGCGGCAACAGCCGUGGCCACAGCCACCAAUGCCACCAAUGC ----------...........((((((.((......)).(---((((........((((((((((.(((((((...)))))))....))))))))))..)))))...)).))))..... ( -45.50, z-score = -1.30, R) >droAna3.scaffold_13337 14142795 91 - 23293914 -------------GCAAUGGAACCGGAACCGGAAGCGGAG---UGGCCAACAACCUGGCCAUGGUGGCGGCCUCGAGUGCGGCUACCGCUGUGGCCACCGCCAGUGG------------ -------------.((.((...(((....)))..((((.(---(.....))....((((((((((((..(((........)))..)))))))))))))))))).)).------------ ( -41.40, z-score = -1.00, R) >droWil1.scaffold_180698 413836 104 - 11422946 GUGGGAAGCGGAGGAAUAACUGCUGCUGCUGGUGAUGGAAGUAUGGCCAAUAACUUAGCCAUUGUGGCGGCUUCAAAUGCGGCAACAGCUGUGGCUACAACUGG--------------- (..(..(((((........))))).)..)((((.(((....))).))))......(((((((.((.((.((.......)).))....)).))))))).......--------------- ( -31.70, z-score = -0.07, R) >droVir3.scaffold_13049 15095791 101 + 25233164 -------------GCUGCGCAACUGC--CUGCCGUCGGAG---UAGCCAACAAUCUAGCCAUGGUGGCUGCCUCCAGUGCAGCAACAGCGGUGGCCACCUCCAGCAGUGCUGCACAGGA -------------((.(.((....))--).))....((((---((((((.((.........)).)))))).)))).((((((((...((((.((...)).)).))..)))))))).... ( -38.80, z-score = 0.74, R) >droMoj3.scaffold_6680 2975860 101 - 24764193 -------------GCUGCACAGAUGC--CAGCUGGCGGGG---UUGCCAACAACUUGGCCAUGGUUGCUGCCUCGAGUGCUGCCACAGCUGUAGCUACCUCCGGGAGUGCUGGGCAGGA -------------.((((.(((..((--((..((((.(((---(((....)))))).)))))))).(((.(((.(((.(((((.......)))))...))).)))))).))).)))).. ( -40.90, z-score = 0.98, R) >consensus __________ACGGAAAUGGAGGUGCAACCGGAGGAGGAG___UGGCCAACAACCUGGCCAUGGUGGCCGCAUCCAGUGCGGCAACAGCCGUGGCCACCGCCACCAAUGCCAGUAAUGC .....................((((...........((........)).......((((((((((.(((((.......)))))....))))))))))....)))).............. (-25.85 = -26.10 + 0.25)

| Location | 2,948,662 – 2,948,768 |
|---|---|
| Length | 106 |
| Sequences | 9 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 70.39 |
| Shannon entropy | 0.58609 |
| G+C content | 0.62468 |
| Mean single sequence MFE | -42.19 |
| Consensus MFE | -21.95 |
| Energy contribution | -22.24 |
| Covariance contribution | 0.30 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.01 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.733821 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 2948662 106 - 24543557 GCAUUACUGGCAUUGGUGGCGGUGGCCACGGCUGUUGCCGCACUUGAUGCGGCCACCAUGGCCAGGUUGUUGGCCA---CUCCUCCUCCGGUUGCACCUCCAUUUCCGU---------- (((..(((((....((.((.(((((((..(((....)))(((.....)))))))))).(((((((....)))))))---..)).)).))))))))..............---------- ( -46.50, z-score = -1.69, R) >droSim1.chr3L 2488377 109 - 22553184 GCAUUACUGGCAUUGGUGGCGGUGGCCACGGCUGUUGCCGCACUGGAUGCGGCCACCAUGGCCAGGUUGUUGGCCACUCCUCCUCCUCCGGUUGCACCUCCAUUUCCGU---------- (((..(((((....((.((.(((((((..(((....)))(((.....)))))))))).(((((((....)))))))..)).))....))))))))..............---------- ( -47.50, z-score = -1.42, R) >droSec1.super_2 2960073 109 - 7591821 GCAUUACUGGCAUUGGUGGCGGUGGCCACGGCUGUUGCCGCACUGGAUGCGGCCACCAUGGCCAGGUUGUUGGCCACUCCUCCUCCUCCGAUUGCUCCUCCAUUUCCGU---------- ........((((((((.((.(((((((..(((....)))(((.....)))))))))).(((((((....)))))))........)).)))).)))).............---------- ( -44.10, z-score = -1.09, R) >droYak2.chr3L 15210655 109 + 24197627 GCAUUACUGGCAUUGGUGGCUGUGGCCACGGCUGUUGCCGCACUGGAUGCGGCCACCAUGGCCAGGUUGUUGGCCACUCCUCCUCCUCCGGUCGCACCUCCAUUUCCGU---------- ((....(((((..((((((((((..(((((((....))))...)))..))))))))))..)))))......((((..............))))))..............---------- ( -45.64, z-score = -1.33, R) >droEre2.scaffold_4784 2970894 106 - 25762168 GCAUUGGUGGCAUUGGUGGCUGUGGCCACGGCUGUUGCCGCACUGGAUGCGGCCACCAUGGCCAGGUUGUUGGCCA---CUCCUCCUCCGGUUGCACCUCCAUUUCCGU---------- (((((((.((...((((((((((..(((((((....))))...)))..))))))))))(((((((....)))))))---.....)).)))).)))..............---------- ( -48.20, z-score = -1.32, R) >droAna3.scaffold_13337 14142795 91 + 23293914 ------------CCACUGGCGGUGGCCACAGCGGUAGCCGCACUCGAGGCCGCCACCAUGGCCAGGUUGUUGGCCA---CUCCGCUUCCGGUUCCGGUUCCAUUGC------------- ------------....(((.(((((((.(.((((...))))....).))))))).)))(((((((....)))))))---........(((....))).........------------- ( -36.40, z-score = -0.14, R) >droWil1.scaffold_180698 413836 104 + 11422946 ---------------CCAGUUGUAGCCACAGCUGUUGCCGCAUUUGAAGCCGCCACAAUGGCUAAGUUAUUGGCCAUACUUCCAUCACCAGCAGCAGCAGUUAUUCCUCCGCUUCCCAC ---------------..((..(((((....((((((((.((.......)).......((((((((....)))))))).............)))))))).)))))..))........... ( -27.10, z-score = -1.85, R) >droVir3.scaffold_13049 15095791 101 - 25233164 UCCUGUGCAGCACUGCUGGAGGUGGCCACCGCUGUUGCUGCACUGGAGGCAGCCACCAUGGCUAGAUUGUUGGCUA---CUCCGACGGCAG--GCAGUUGCGCAGC------------- ..(((((((((.((((((..((.((((.(((.(((....))).))).)))(((((.((.........)).))))).---).))..))))))--...))))))))).------------- ( -47.30, z-score = -0.96, R) >droMoj3.scaffold_6680 2975860 101 + 24764193 UCCUGCCCAGCACUCCCGGAGGUAGCUACAGCUGUGGCAGCACUCGAGGCAGCAACCAUGGCCAAGUUGUUGGCAA---CCCCGCCAGCUG--GCAUCUGUGCAGC------------- ..((((.(((..((.((.(((((.(((((....))))).)).)))..)).))........((((....((((((..---....))))))))--))..))).)))).------------- ( -37.00, z-score = 0.74, R) >consensus GCAUUACUGGCAUUGGUGGCGGUGGCCACGGCUGUUGCCGCACUGGAUGCGGCCACCAUGGCCAGGUUGUUGGCCA___CUCCUCCUCCGGUUGCACCUCCAUUUCCGU__________ .................((.(((((((...((....)).((.......))))))))).(((((((....))))))).....)).................................... (-21.95 = -22.24 + 0.30)
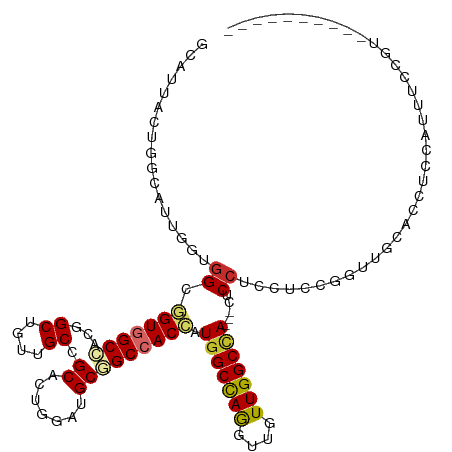
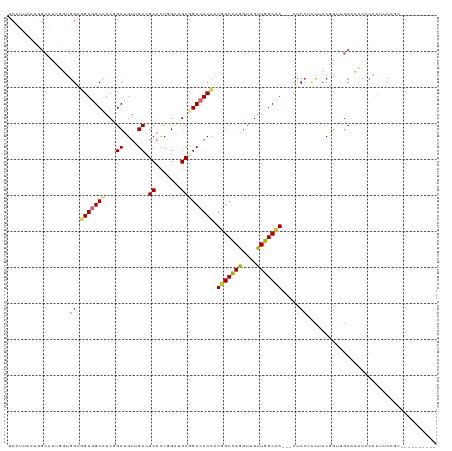
| Location | 2,948,688 – 2,948,794 |
|---|---|
| Length | 106 |
| Sequences | 9 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 67.81 |
| Shannon entropy | 0.62614 |
| G+C content | 0.64060 |
| Mean single sequence MFE | -43.53 |
| Consensus MFE | -25.46 |
| Energy contribution | -26.26 |
| Covariance contribution | 0.79 |
| Combinations/Pair | 1.26 |
| Mean z-score | -0.93 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.910635 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
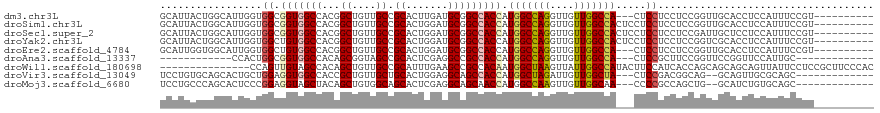
>dm3.chr3L 2948688 106 + 24543557 GGAGUGGCCAACAACCUGGCCAUGGUGGCCGCAUCAAGUGCGGCAACAGCCGUGGCCACCGCCACCAAUGCCAGUAAUGCCAGCAAUACCGCCCCCGGGUCAGAGG--------------- ((.(((((........((((((((((.(((((((...)))))))....))))))))))..)))))...(((((....))...)))...))(((....)))......--------------- ( -43.60, z-score = -0.92, R) >droSim1.chr3L 2488406 106 + 22553184 GGAGUGGCCAACAACCUGGCCAUGGUGGCCGCAUCCAGUGCGGCAACAGCCGUGGCCACCGCCACCAAUGCCAGUAAUGCCAGCAAUACCGCCCCCGGGUCAGAGG--------------- ((.(((((........((((((((((.(((((((...)))))))....))))))))))..)))))...(((((....))...)))...))(((....)))......--------------- ( -43.60, z-score = -0.85, R) >droSec1.super_2 2960102 106 + 7591821 GGAGUGGCCAACAACCUGGCCAUGGUGGCCGCAUCCAGUGCGGCAACAGCCGUGGCCACCGCCACCAAUGCCAGUAAUGCCAUCAAUACCGCCCCCGGGUCAGAGG--------------- ((.(((((........((((((((((.(((((((...)))))))....))))))))))..)))))..(((.((....)).))).....))(((....)))......--------------- ( -43.40, z-score = -1.04, R) >droYak2.chr3L 15210684 106 - 24197627 GGAGUGGCCAACAACCUGGCCAUGGUGGCCGCAUCCAGUGCGGCAACAGCCGUGGCCACAGCCACCAAUGCCAGUAAUGCCAGUAACACCGCUCCCGGAACGGAGG--------------- ((.(((((........((((((((((.(((((((...)))))))....))))))))))..)))))...((.((....)).))......)).((((......)))).--------------- ( -44.80, z-score = -1.55, R) >droEre2.scaffold_4784 2970920 106 + 25762168 GGAGUGGCCAACAACCUGGCCAUGGUGGCCGCAUCCAGUGCGGCAACAGCCGUGGCCACAGCCACCAAUGCCACCAAUGCCAGUAACACCGCCCCCGGGUCAGAGG--------------- ((.(.(((.......(((((..(((((((.((((...))))(((....)))(((((....)))))....)))))))..))))).......))))))..........--------------- ( -43.04, z-score = -0.84, R) >droAna3.scaffold_13337 14142818 94 - 23293914 GGAGUGGCCAACAACCUGGCCAUGGUGGCGGCCUCGAGUGCGGCUACCGCUGUGGCCACCGCCAGUGGCA----GCAACUCCGCAGCUC-----CAGGAGCUG------------------ (((((.((((......((((((((((((..(((........)))..)))))))))))).......)))).----...))))).((((((-----...))))))------------------ ( -50.42, z-score = -2.28, R) >droWil1.scaffold_180698 413875 85 - 11422946 AGUAUGGCCAAUAACUUAGCCAUUGUGGCGGCUUCAAAUGCGGCAACAGCUGUGGCUACAAC---UGGCA----GCAACAGCGCA---------CAGGAGG-------------------- ....((((((......((((..((((.(((........))).))))..))))))))))(..(---((((.----((....)))).---------)))..).-------------------- ( -25.30, z-score = 0.52, R) >droVir3.scaffold_13049 15095812 109 + 25233164 GGAGUAGCCAACAAUCUAGCCAUGGUGGCUGCCUCCAGUGCAGCAACAGCGGUGGCCACCUCCAGCAGUGCUGCACAGGAUAGCGG---------AGGAGCGGCCUCCUCGUCG--CAAG- ((((((((((.((.........)).)))))).)))).((((((((...((((.((...)).)).))..))))))))......((((---------(((((....))))))..))--)...- ( -47.90, z-score = -1.69, R) >droMoj3.scaffold_6680 2975881 118 - 24764193 GGGGUUGCCAACAACUUGGCCAUGGUUGCUGCCUCGAGUGCUGCCACAGCUGUAGCUACCUCCGGGAGUGCUGGGCAGGAGAACGGCGAGG---CAGGAGCAGCUGCUGCCUCCACCAAAG (((((.(((((....)))))....(((.((((((((.((.(((((.((((....(((.((....))))))))))))))....))..)))))---))).))).......))).))....... ( -49.70, z-score = 0.23, R) >consensus GGAGUGGCCAACAACCUGGCCAUGGUGGCCGCAUCCAGUGCGGCAACAGCCGUGGCCACCGCCACCAAUGCCAGUAAUGCCAGCAACACCGC_CCCGGAGCAGAGG_______________ ((.(((((((......)))))))(((((((((.....))(((((....)))))))))))).)).......................................................... (-25.46 = -26.26 + 0.79)
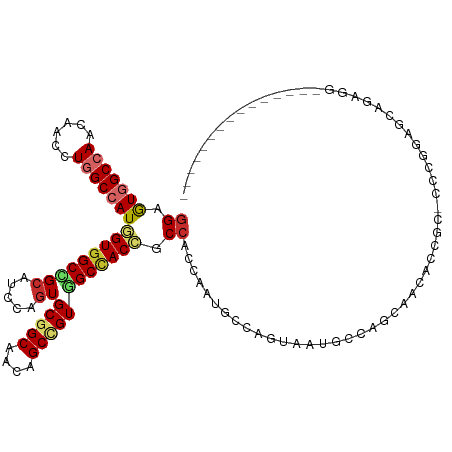
| Location | 2,948,688 – 2,948,794 |
|---|---|
| Length | 106 |
| Sequences | 9 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 67.81 |
| Shannon entropy | 0.62614 |
| G+C content | 0.64060 |
| Mean single sequence MFE | -46.69 |
| Consensus MFE | -22.20 |
| Energy contribution | -22.60 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.36 |
| Mean z-score | -0.95 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.687493 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 2948688 106 - 24543557 ---------------CCUCUGACCCGGGGGCGGUAUUGCUGGCAUUACUGGCAUUGGUGGCGGUGGCCACGGCUGUUGCCGCACUUGAUGCGGCCACCAUGGCCAGGUUGUUGGCCACUCC ---------------((((((...)))))).((....((..(((...(((((..(((((((((..((....))..))..((((.....)))))))))))..)))))..)))..))....)) ( -48.80, z-score = -0.74, R) >droSim1.chr3L 2488406 106 - 22553184 ---------------CCUCUGACCCGGGGGCGGUAUUGCUGGCAUUACUGGCAUUGGUGGCGGUGGCCACGGCUGUUGCCGCACUGGAUGCGGCCACCAUGGCCAGGUUGUUGGCCACUCC ---------------((((((...)))))).((....((..(((...(((((..(((((((.((..(((((((....))))...)))..)).)))))))..)))))..)))..))....)) ( -53.00, z-score = -1.57, R) >droSec1.super_2 2960102 106 - 7591821 ---------------CCUCUGACCCGGGGGCGGUAUUGAUGGCAUUACUGGCAUUGGUGGCGGUGGCCACGGCUGUUGCCGCACUGGAUGCGGCCACCAUGGCCAGGUUGUUGGCCACUCC ---------------((((((...)))))).(.(((..(((.((....)).)))..))).)(((((((..(((....)))(((.....)))))))))).(((((((....))))))).... ( -49.40, z-score = -0.89, R) >droYak2.chr3L 15210684 106 + 24197627 ---------------CCUCCGUUCCGGGAGCGGUGUUACUGGCAUUACUGGCAUUGGUGGCUGUGGCCACGGCUGUUGCCGCACUGGAUGCGGCCACCAUGGCCAGGUUGUUGGCCACUCC ---------------.((((......)))).((.((..(..(((...(((((..((((((((((..(((((((....))))...)))..))))))))))..)))))..)))..)..)).)) ( -53.50, z-score = -2.38, R) >droEre2.scaffold_4784 2970920 106 - 25762168 ---------------CCUCUGACCCGGGGGCGGUGUUACUGGCAUUGGUGGCAUUGGUGGCUGUGGCCACGGCUGUUGCCGCACUGGAUGCGGCCACCAUGGCCAGGUUGUUGGCCACUCC ---------------((((((...)))))).(((((((((......)))))))))(((((((((..(((((((....))))...)))..))))))))).(((((((....))))))).... ( -51.20, z-score = -0.86, R) >droAna3.scaffold_13337 14142818 94 + 23293914 ------------------CAGCUCCUG-----GAGCUGCGGAGUUGC----UGCCACUGGCGGUGGCCACAGCGGUAGCCGCACUCGAGGCCGCCACCAUGGCCAGGUUGUUGGCCACUCC ------------------((((((...-----)))))).(((((.((----(((((.(((.(((((((.(.((((...))))....).))))))).))))))).........))).))))) ( -45.00, z-score = -0.69, R) >droWil1.scaffold_180698 413875 85 + 11422946 --------------------CCUCCUG---------UGCGCUGUUGC----UGCCA---GUUGUAGCCACAGCUGUUGCCGCAUUUGAAGCCGCCACAAUGGCUAAGUUAUUGGCCAUACU --------------------.....((---------((.(((((.((----(((..---...))))).)))))....((.((.......)).))))))((((((((....))))))))... ( -30.00, z-score = -1.64, R) >droVir3.scaffold_13049 15095812 109 - 25233164 -CUUG--CGACGAGGAGGCCGCUCCU---------CCGCUAUCCUGUGCAGCACUGCUGGAGGUGGCCACCGCUGUUGCUGCACUGGAGGCAGCCACCAUGGCUAGAUUGUUGGCUACUCC -....--(((((((((((.....)))---------))(((.(((.(((((((((.((.((.((...)).)))).).)))))))).))))))((((.....))))...))))))........ ( -45.60, z-score = -0.71, R) >droMoj3.scaffold_6680 2975881 118 + 24764193 CUUUGGUGGAGGCAGCAGCUGCUCCUG---CCUCGCCGUUCUCCUGCCCAGCACUCCCGGAGGUAGCUACAGCUGUGGCAGCACUCGAGGCAGCAACCAUGGCCAAGUUGUUGGCAACCCC ...((((((((.(((...)))))))((---(((((..((....(((((((((.(((...)))((....)).)))).))))).)).)))))))...))))..(((((....)))))...... ( -43.70, z-score = 0.91, R) >consensus _______________CCUCUGAUCCGGG_GCGGUGUUGCUGGCAUUACUGGCAUUGGUGGCGGUGGCCACGGCUGUUGCCGCACUGGAUGCGGCCACCAUGGCCAGGUUGUUGGCCACUCC ..........................................................((.(((((((((((((((.(((((.......))))).))........))))).)))))))))) (-22.20 = -22.60 + 0.40)
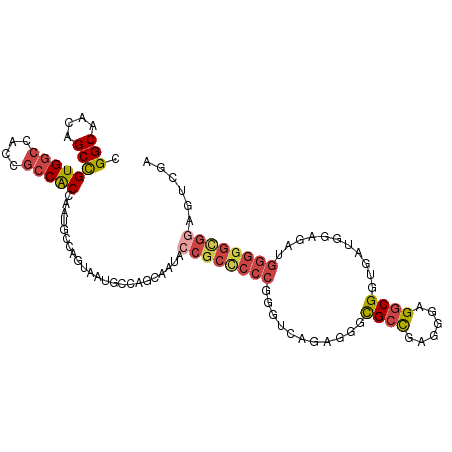
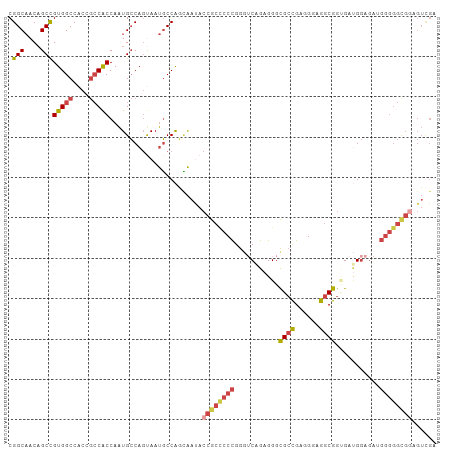
| Location | 2,948,728 – 2,948,834 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 77.61 |
| Shannon entropy | 0.44132 |
| G+C content | 0.68291 |
| Mean single sequence MFE | -46.70 |
| Consensus MFE | -28.34 |
| Energy contribution | -29.57 |
| Covariance contribution | 1.23 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.23 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.596934 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 2948728 106 + 24543557 CGGCAACAGCCGUGGCCACCGCCACCAAUGCCAGUAAUGCCAGCAAUACCGCCCCCGGGUCAGAGGGCGCCGAGGGAGGCGGUGAUGGAGAUGGGGGCGGUGUCGA ((((....))))((((.((((((.(((...(((....((....)).(((((((((((((((....))).))..))).))))))).)))...))).)))))))))). ( -50.80, z-score = -1.44, R) >droSim1.chr3L 2488446 106 + 22553184 CGGCAACAGCCGUGGCCACCGCCACCAAUGCCAGUAAUGCCAGCAAUACCGCCCCCGGGUCAGAGGGCGCCGAGGGAGGCGGUGAUGGAGAAGGGGGCGGAGUCGA ((((....))))((((..(((((.((....(((....((....)).(((((((((((((((....))).))..))).))))))).))).....))))))).)))). ( -46.90, z-score = -0.95, R) >droSec1.super_2 2960142 106 + 7591821 CGGCAACAGCCGUGGCCACCGCCACCAAUGCCAGUAAUGCCAUCAAUACCGCCCCCGGGUCAGAGGGCGCCGAGGGAGGCGGUGAUGGAGAAGGGGGCGGAGUCGA ((((....))))((((..(((((.((.....((....))(((((...((((((((((((((....))).))..))).)))))))))))....)).))))).)))). ( -48.80, z-score = -1.57, R) >droYak2.chr3L 15210724 106 - 24197627 CGGCAACAGCCGUGGCCACAGCCACCAAUGCCAGUAAUGCCAGUAACACCGCUCCCGGAACGGAGGGCGCCGAGGGAGGCGGCGAUGGAGAUGGGGGUGUGGUUGA ((((....)))).(((((((.((.(((...(((....((((.........(((((((...))).))))(((......))))))).)))...))))).))))))).. ( -47.60, z-score = -1.63, R) >droEre2.scaffold_4784 2970960 106 + 25762168 CGGCAACAGCCGUGGCCACAGCCACCAAUGCCACCAAUGCCAGUAACACCGCCCCCGGGUCAGAGGGCGCCGAGGGAGGCGGAGACGGAGAUGGGGGCGGGGUUGA .((((...((.(((((....)))))....))......))))..((((.(((((((((..((.......(((......)))(....).))..))))))))).)))). ( -46.60, z-score = -0.75, R) >droAna3.scaffold_13337 14142858 85 - 23293914 CGGCUACCGCUGUGGCCAC---CGCCAGUGGCAGCAACUCCGCAGCUCCAGGAGCUGGAGCAGAGGGUGGUGUCGAAGGCGAAGGGGA------------------ .((((((....))))))..---((((..(((((.((.(.((...(((((((...)))))))...))))).)))))..)))).......------------------ ( -39.50, z-score = -1.06, R) >consensus CGGCAACAGCCGUGGCCACCGCCACCAAUGCCAGUAAUGCCAGCAAUACCGCCCCCGGGUCAGAGGGCGCCGAGGGAGGCGGUGAUGGAGAUGGGGGCGGAGUCGA .(((....)))(((((....))))).......................((((((((...........((((......))))...........))))))))...... (-28.34 = -29.57 + 1.23)

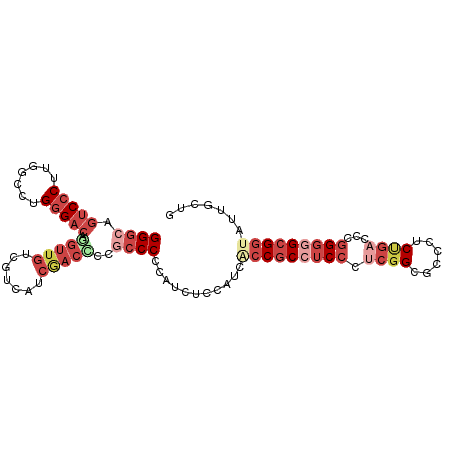
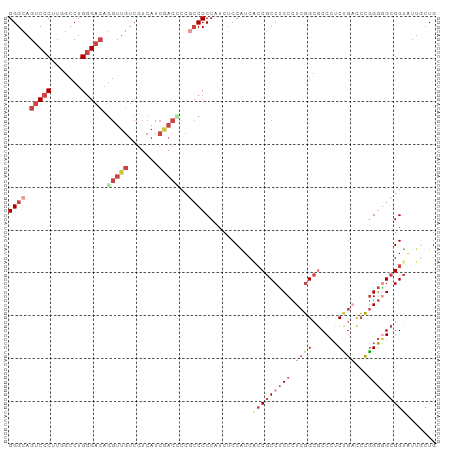
| Location | 2,948,768 – 2,948,868 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 83.00 |
| Shannon entropy | 0.33291 |
| G+C content | 0.67456 |
| Mean single sequence MFE | -40.22 |
| Consensus MFE | -23.97 |
| Energy contribution | -27.42 |
| Covariance contribution | 3.45 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr3L 2948768 100 - 24543557 GGGCAGUCCCUUGGCCUGGGAAAGGUUGUCGUCAUCGACACCGCCCCCAUCUCCAUCACCGCCUCCCUCGGCGCCCUCUGACCCGGGGGCGGUAUUGCUG ((((..((((.......))))..(((.((((....))))))))))).......((..((((((......)))((((((......)))))))))..))... ( -42.70, z-score = -1.46, R) >droSim1.chr3L 2488486 100 - 22553184 GGGCAGUCCCUUGGCCUGGGACAGGUUGUCGUCAUCGACUCCGCCCCCUUCUCCAUCACCGCCUCCCUCGGCGCCCUCUGACCCGGGGGCGGUAUUGCUG ((((.(((((.......))))).((..((((....)))).)))))).......((..((((((......)))((((((......)))))))))..))... ( -44.60, z-score = -2.00, R) >droSec1.super_2 2960182 100 - 7591821 GGGCAGUCCCUUGGCCUGGGACAAGUUGUCGUCAUCGACUCCGCCCCCUUCUCCAUCACCGCCUCCCUCGGCGCCCUCUGACCCGGGGGCGGUAUUGAUG ((((.(((((.......)))))..(..((((....))))..))))).......(((((..(((.(((((((.(........)))))))).)))..))))) ( -44.30, z-score = -2.57, R) >droYak2.chr3L 15210764 100 + 24197627 GGGCAGUCCCUUCGCCUGGGACAGGUUGUCGUCAUCAACCACACCCCCAUCUCCAUCGCCGCCUCCCUCGGCGCCCUCCGUUCCGGGAGCGGUGUUACUG (((..(((((.......))))).(((((.......))))).....)))........((((((.((((..((((.....))))..))))))))))...... ( -39.00, z-score = -2.50, R) >droEre2.scaffold_4784 2971000 100 - 25762168 GGGCAGUCCCUUCGCCUGGGACAGGUUGUCGUCAUCAACCCCGCCCCCAUCUCCGUCUCCGCCUCCCUCGGCGCCCUCUGACCCGGGGGCGGUGUUACUG ((((.(((((.......))))).(((((.......)))))..)))).((((.........(((......)))((((((......))))))))))...... ( -41.60, z-score = -1.82, R) >droAna3.scaffold_13337 14142886 91 + 23293914 GGGCAGGCCCUUUGCCUGUGACAGGUUGUCCUCAU---------CCCCUUCGCCUUCGACACCACCCUCUGCUCCAGCUCCUGGAGCUGCGGAGUUGCUG ((((((((.....))))))(((.....))).....---------.))((((((....((........)).(((((((...))))))).))))))...... ( -29.10, z-score = 0.04, R) >consensus GGGCAGUCCCUUGGCCUGGGACAGGUUGUCGUCAUCGACCCCGCCCCCAUCUCCAUCACCGCCUCCCUCGGCGCCCUCUGACCCGGGGGCGGUAUUGCUG ((((.(((((.......))))).(((((.......)))))..))))...........(((((((((.((((......))))...)))))))))....... (-23.97 = -27.42 + 3.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:59:15 2011