
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 2,942,038 – 2,942,139 |
| Length | 101 |
| Max. P | 0.725890 |

| Location | 2,942,038 – 2,942,139 |
|---|---|
| Length | 101 |
| Sequences | 8 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 67.54 |
| Shannon entropy | 0.67865 |
| G+C content | 0.41632 |
| Mean single sequence MFE | -28.28 |
| Consensus MFE | -5.39 |
| Energy contribution | -6.25 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.54 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.19 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.725890 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 2942038 101 - 24543557 CAGCAGUCGCAGGACUUACG--UCCGACUUCUGUG-AUUGCUAUUUACACGCCUCU-AGGCUGG-AUUUUAGAUACAGCUUGGUAACGGAACGAAAGUUAAACCAA .(((((((((((((.((...--...)).)))))))-)))))).......((...((-((((((.-..........))))))))...)).(((....)))....... ( -33.30, z-score = -3.40, R) >droAna3.scaffold_13337 14136603 103 + 23293914 CUCCAGUUGCCGGACACGGG-AUCCGGCUUCUGUGAAAUGCUAUUUACACGCCUCU-UGGCUGGAAUUAUUCAACCAACUUGA-AACAAAGAUAUAGAAGAACCAA .(((((((((((((......-.))))))...((((((......)))))).......-.)))))))....(((((.....))))-)..................... ( -23.30, z-score = -0.85, R) >droEre2.scaffold_4784 2964219 101 - 25762168 CAGCAGUCGCAGGACUUGCG--UCCGACUUCUGUG-AUUGCUAUUUACACGCCUCU-AGGCUGG-AUUUUAGAUACAGCUUGGUAACGGAACGAAAGUUAAACCAA .(((((((((((((.(((..--..))).)))))))-)))))).......((...((-((((((.-..........))))))))...)).(((....)))....... ( -34.90, z-score = -3.47, R) >droYak2.chr3L 15203831 101 + 24197627 CAGCAGUCGCAGGACUUGCG--UCCGACUUCUGUG-AUUGCUAUUUACACGCCUCU-AGGCUGG-AUUUUAGAUACAGCUUGGUAACGGAACGAAAGUUAAACCAA .(((((((((((((.(((..--..))).)))))))-)))))).......((...((-((((((.-..........))))))))...)).(((....)))....... ( -34.90, z-score = -3.47, R) >droSec1.super_2 2953512 101 - 7591821 CAGCAGUCGCAGGACUCGCG--UCCGACUUCUGUG-AUUGCUAUUUACACGCCUCU-AGGCUGG-AUUUUAGAUACAGCUUGGUAACGGAACGAAAGUUAAACCAA .(((((((((((((.(((..--..))).)))))))-)))))).......((...((-((((((.-..........))))))))...)).(((....)))....... ( -37.00, z-score = -4.01, R) >droSim1.chr3L 2481777 101 - 22553184 CAGCAGUCGCAGGACUCGCG--UCCGACUUCUGUG-AUUGCUAUUUACACGCCUCU-AGGCUGG-AUUUUAGAUACAGCUUGGUAACGGAACGAAAGUUAAACCAA .(((((((((((((.(((..--..))).)))))))-)))))).......((...((-((((((.-..........))))))))...)).(((....)))....... ( -37.00, z-score = -4.01, R) >droWil1.scaffold_180698 405585 100 + 11422946 ------UAGCAGUCGAUGAGGUGUCUUUAUUGAUCAAUAUGUAUUUACACGCAUCUUUAAUUAAAUUCUUUUCUGGUUUUUUUAAACUUGAAAUUAUGUAAGUAAA ------.....((((((((((...)))))))))).......((((((((..((..(((((..(((((.......)))))..)))))..))......)))))))).. ( -14.90, z-score = -0.29, R) >droVir3.scaffold_13049 15088218 87 - 25233164 --------AAAUAAUUUAAGAAAUCGACUAGUUAACGCAACUAUUUACACGCCUUC---GUUAG-ACAGUUGAAACA-------AAUGAAUUGAAAUAGAAAUCAA --------...(((((((.....((((((..((((((..................)---)))))-..))))))....-------..)))))))............. ( -10.97, z-score = -0.66, R) >consensus CAGCAGUCGCAGGACUUGCG__UCCGACUUCUGUG_AUUGCUAUUUACACGCCUCU_AGGCUGG_AUUUUAGAUACAGCUUGGUAACGGAACGAAAGUUAAACCAA .......(((((((.(((......))).)))))))...............(((.....)))............................................. ( -5.39 = -6.25 + 0.86)

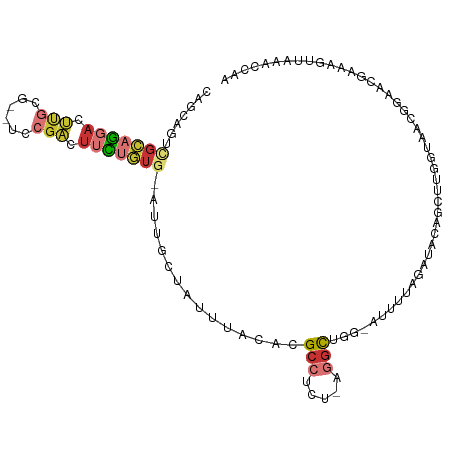

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:59:10 2011