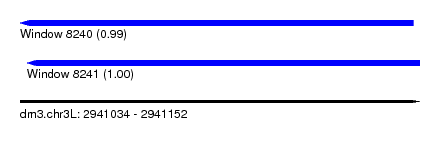
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 2,941,034 – 2,941,152 |
| Length | 118 |
| Max. P | 0.998527 |

| Location | 2,941,034 – 2,941,150 |
|---|---|
| Length | 116 |
| Sequences | 14 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 73.01 |
| Shannon entropy | 0.62248 |
| G+C content | 0.52504 |
| Mean single sequence MFE | -37.09 |
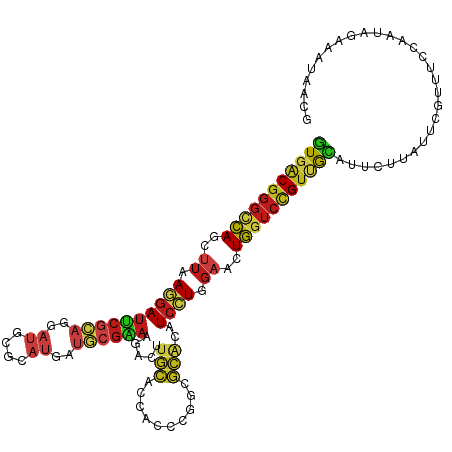
| Consensus MFE | -19.66 |
| Energy contribution | -19.96 |
| Covariance contribution | 0.29 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.992074 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
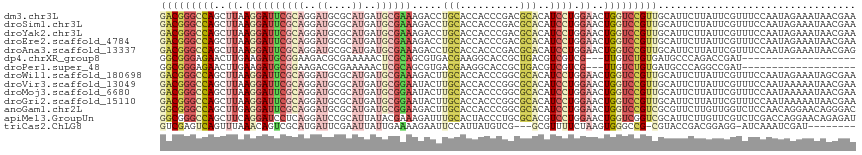
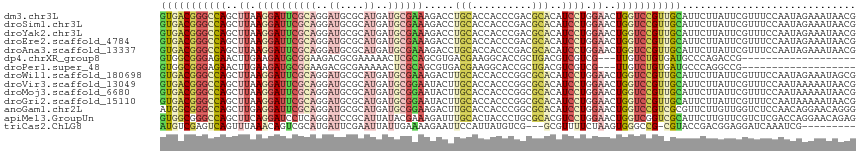
>dm3.chr3L 2941034 116 - 24543557 GACGGGCCAGCUUAAGGAUUCGCAGGAUGCGCAUGAUGCGAAAGACCUGCACCACCCGACGCACAUCCUGGAACUGGUCCGUUGCAUUCUUAUUCGUUUCCAAUAGAAAUAACGAA ((((((((((.((.(((((..(((((...(((.....))).....))))).......(.....)))))).)).)))))))))).........((((((............)))))) ( -36.50, z-score = -2.21, R) >droSim1.chr3L 2480737 116 - 22553184 GACGGGCCAGCUUAAGGAUUCGCAGGAUGCGCAUGAUGCGAAAGACCUGCACCACCCGACGCACAUCCUGGAACUGGUCCGUUGCAUUCUUAUUCGUUUCCAAUAGAAAUAACGAA ((((((((((.((.(((((..(((((...(((.....))).....))))).......(.....)))))).)).)))))))))).........((((((............)))))) ( -36.50, z-score = -2.21, R) >droYak2.chr3L 15202809 116 + 24197627 GACGGGCCAGCUUAAGGAUUCGCAGGAUGCGCAUGAUGCGAAAGACCUGCACCACCCGACGCACAUCCUGGAACUGGUCCGUUGCAUUCUUAUUCGUUUCCAAUAGAAAUAACGAA ((((((((((.((.(((((..(((((...(((.....))).....))))).......(.....)))))).)).)))))))))).........((((((............)))))) ( -36.50, z-score = -2.21, R) >droEre2.scaffold_4784 2963227 116 - 25762168 GACGGGCCAGCUUAAGGAUUCGCAGGAUGCGCAUGAUGCGAAAGACCUGCACCACCCGACGCACAUCCUGGAACUGGUCCGUUGCAUUCUUAUUCGUUUCCAAUAGAAAUAACGAA ((((((((((.((.(((((..(((((...(((.....))).....))))).......(.....)))))).)).)))))))))).........((((((............)))))) ( -36.50, z-score = -2.21, R) >droAna3.scaffold_13337 14135613 116 + 23293914 GACGGGCCAGCUUAAGGAUUCGCAGGAUGCGCAUGAUGCGAAAGACCUGCACCACCCGACGCACAUCCUGGAACUGGUCCGUUGCAUUCUUAUUCGUUUCCAAUAGAAAUAACGAG ((((((((((.((.(((((..(((((...(((.....))).....))))).......(.....)))))).)).))))))))))..........(((((............))))). ( -36.30, z-score = -2.21, R) >dp4.chrXR_group8 1570099 94 + 9212921 GGCGGGAGAACUUGAAGAUGCGGAAGACGCGAAAAACUCGCAGCGUGACGAAGGCACCGCUGACGUCGUCG---UUGUCUGUGAUGCCCAGACCGAU------------------- ((((((............((((.....))))....((..(((((((((((..(((...)))..))))).))---))))..))....))).).))...------------------- ( -28.50, z-score = 0.23, R) >droPer1.super_48 160285 94 + 581423 GGCGGGAGAACUUGAAGAUGCGGAAGACGCGAAAAACUCGCAGCGUGACGAAGGCACCGCUGACGUCGUCG---UUGUCUGUGAUGCCCAGGCCGAU------------------- ((((((............((((.....))))....((..(((((((((((..(((...)))..))))).))---))))..))....)))..)))...------------------- ( -30.90, z-score = -0.04, R) >droWil1.scaffold_180698 404620 116 + 11422946 GACGGGCCAGCUUAAGGAUUCGCAGGAUGCGCAUGAUGCGAAAGACUUGCACCACCCGGCGCACAUCCUGGAACUGGUCCGUUGCAUUCUUAUUCGUUUCCAAUAGAAAUAGCGAA ((((((((((........(((.((((((((((.((.(((((.....))))).))....)))))..)))))))))))))))))).........((((((............)))))) ( -39.80, z-score = -2.18, R) >droVir3.scaffold_13049 15087365 116 - 25233164 GACGGGCCAGCUUAAGGAUUCGCAGGAUGCGCAUGAUGCGGAAUACUUGCACCACCCGGCGCACAUCCUGGAACUGGUCCGUUGCAUUCUUAUUCGUUUCCAAUAAAAAUAACGAA ((((((((((........(((.((((((((((.....))).......(((.((....)).))))))))))))))))))))))).........((((((............)))))) ( -39.40, z-score = -2.58, R) >droMoj3.scaffold_6680 2967293 116 + 24764193 GACGGGCCAGCUUAAGGAUUCGCAGGAUGCGCAUGAUGCGGAAUACUUGCACCACCCGGCGCACAUCCUGGAACUGGUCCGUUGCAUUCUUAUUCGUUUCCAAUAAAAAUAACGAA ((((((((((........(((.((((((((((.....))).......(((.((....)).))))))))))))))))))))))).........((((((............)))))) ( -39.40, z-score = -2.58, R) >droGri2.scaffold_15110 19503591 116 - 24565398 GACGGGCCAGCUUAAGGAUUCGCAGGAUGCGCAUGAUGCGGAAUACUUGCACCACCCGGCGCACAUCCUGGAACUGGUCCGUUGCAUUCUUAUUCGUUUCCAAUAAAAAUAACGAA ((((((((((........(((.((((((((((.....))).......(((.((....)).))))))))))))))))))))))).........((((((............)))))) ( -39.40, z-score = -2.58, R) >anoGam1.chr2L 25639249 116 + 48795086 GGCGGGCCAGCUUGAGGAUUCGCAGGAUGCGCAUGAUGCGGAAGACUUGCACCACCCGGCGCACAUCCUGGAACUGGUCCGUCGCGUUCUUGUUGGUCUCCAACAGGAACAGGGAC ((((((((((.((.((((((((((..((....))..)))))).....(((.((....)).)))..)))).)).))))))))))..(((((((((((...)))))))))))...... ( -54.60, z-score = -3.45, R) >apiMel3.GroupUn 83739116 116 - 399230636 GGCGGGCCAGCUUCAGGAUCCUCAGGAUCCGCAUUAUACGAAAGAUUUGCACUACCCUGCGCACGUCCUGGAACUGGUCGGUCGCAUUCUUGUUCGUCUCGACCAGGAACAGAGAU (((.((((((.((((((((.(.((((....(((..((.(....))).))).....)))).)...)))))))).)))))).)))...((((((.(((...))).))))))....... ( -41.30, z-score = -1.78, R) >triCas2.ChLG8 12910646 103 - 15773733 GUCGAGUCAGUUUAAACAGUCGCAUGAUUCGAAUUAUUGAAAAGAAUUCCAUUAUGUCG---GCGUUUUCUAAGUGGGCCG-CGUACCGACGGAGG-AUCAAAUCGAU-------- (((((.............((((((((((..(((((.((....))))))).)))))).))---))(((((((...(((....-....)))..)))))-))....)))))-------- ( -23.70, z-score = 0.23, R) >consensus GACGGGCCAGCUUAAGGAUUCGCAGGAUGCGCAUGAUGCGAAAGACUUGCACCACCCGGCGCACAUCCUGGAACUGGUCCGUUGCAUUCUUAUUCGUUUCCAAUAGAAAUAACGAA (((((((((..((.((((((((((..((....))..)))))).....(((..........)))..)))).))..)))))))))................................. (-19.66 = -19.96 + 0.29)
| Location | 2,941,036 – 2,941,152 |
|---|---|
| Length | 116 |
| Sequences | 14 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 73.06 |
| Shannon entropy | 0.61895 |
| G+C content | 0.53148 |
| Mean single sequence MFE | -38.81 |
| Consensus MFE | -22.92 |
| Energy contribution | -22.95 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.54 |
| Mean z-score | -2.19 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.39 |
| SVM RNA-class probability | 0.998527 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
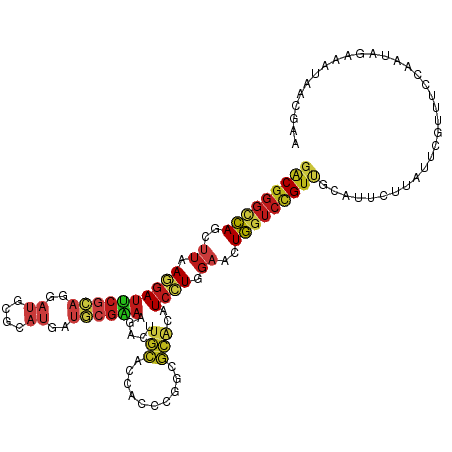
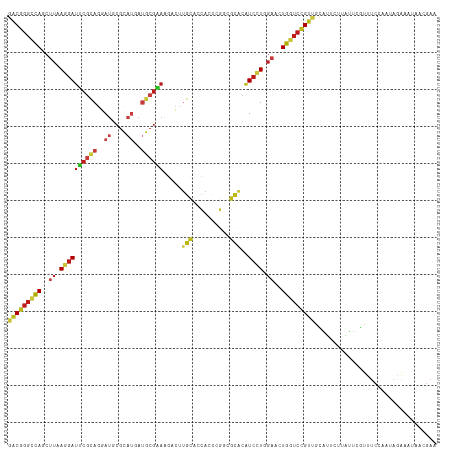
>dm3.chr3L 2941036 116 - 24543557 GUGACGGGCCAGCUUAAGGAUUCGCAGGAUGCGCAUGAUGCGAAAGACCUGCACCACCCGACGCACAUCCUGGAACUGGUCCGUUGCAUUCUUAUUCGUUUCCAAUAGAAAUAACG (..(((((((((.((.(((((..(((((...(((.....))).....))))).......(.....)))))).)).)))))))))..)..........(((((.....))))).... ( -39.40, z-score = -2.90, R) >droSim1.chr3L 2480739 116 - 22553184 GUGACGGGCCAGCUUAAGGAUUCGCAGGAUGCGCAUGAUGCGAAAGACCUGCACCACCCGACGCACAUCCUGGAACUGGUCCGUUGCAUUCUUAUUCGUUUCCAAUAGAAAUAACG (..(((((((((.((.(((((..(((((...(((.....))).....))))).......(.....)))))).)).)))))))))..)..........(((((.....))))).... ( -39.40, z-score = -2.90, R) >droYak2.chr3L 15202811 116 + 24197627 GUGACGGGCCAGCUUAAGGAUUCGCAGGAUGCGCAUGAUGCGAAAGACCUGCACCACCCGACGCACAUCCUGGAACUGGUCCGUUGCAUUCUUAUUCGUUUCCAAUAGAAAUAACG (..(((((((((.((.(((((..(((((...(((.....))).....))))).......(.....)))))).)).)))))))))..)..........(((((.....))))).... ( -39.40, z-score = -2.90, R) >droEre2.scaffold_4784 2963229 116 - 25762168 GUGACGGGCCAGCUUAAGGAUUCGCAGGAUGCGCAUGAUGCGAAAGACCUGCACCACCCGACGCACAUCCUGGAACUGGUCCGUUGCAUUCUUAUUCGUUUCCAAUAGAAAUAACG (..(((((((((.((.(((((..(((((...(((.....))).....))))).......(.....)))))).)).)))))))))..)..........(((((.....))))).... ( -39.40, z-score = -2.90, R) >droAna3.scaffold_13337 14135615 116 + 23293914 GUGACGGGCCAGCUUAAGGAUUCGCAGGAUGCGCAUGAUGCGAAAGACCUGCACCACCCGACGCACAUCCUGGAACUGGUCCGUUGCAUUCUUAUUCGUUUCCAAUAGAAAUAACG (..(((((((((.((.(((((..(((((...(((.....))).....))))).......(.....)))))).)).)))))))))..)..........(((((.....))))).... ( -39.40, z-score = -2.90, R) >dp4.chrXR_group8 1570101 94 + 9212921 GUGGCGGGAGAACUUGAAGAUGCGGAAGACGCGAAAAACUCGCAGCGUGACGAAGGCACCGCUGACGUCGUCG---UUGUCUGUGAUGCCCAGACCG------------------- ..((((((............((((.....))))....((..(((((((((((..(((...)))..))))).))---))))..))....))).).)).------------------- ( -28.50, z-score = 0.52, R) >droPer1.super_48 160287 94 + 581423 GUGGCGGGAGAACUUGAAGAUGCGGAAGACGCGAAAAACUCGCAGCGUGACGAAGGCACCGCUGACGUCGUCG---UUGUCUGUGAUGCCCAGGCCG------------------- ..((((((............((((.....))))....((..(((((((((((..(((...)))..))))).))---))))..))....)))..))).------------------- ( -30.90, z-score = 0.24, R) >droWil1.scaffold_180698 404622 116 + 11422946 GUGACGGGCCAGCUUAAGGAUUCGCAGGAUGCGCAUGAUGCGAAAGACUUGCACCACCCGGCGCACAUCCUGGAACUGGUCCGUUGCAUUCUUAUUCGUUUCCAAUAGAAAUAGCG (..(((((((((........(((.((((((((((.((.(((((.....))))).))....)))))..)))))))))))))))))..)..........(((((.....))))).... ( -42.70, z-score = -2.85, R) >droVir3.scaffold_13049 15087367 116 - 25233164 GUGACGGGCCAGCUUAAGGAUUCGCAGGAUGCGCAUGAUGCGGAAUACUUGCACCACCCGGCGCACAUCCUGGAACUGGUCCGUUGCAUUCUUAUUCGUUUCCAAUAAAAAUAACG (..(((((((((........(((.((((((((((.....))).......(((.((....)).))))))))))))))))))))))..)............................. ( -40.80, z-score = -2.84, R) >droMoj3.scaffold_6680 2967295 116 + 24764193 GUGACGGGCCAGCUUAAGGAUUCGCAGGAUGCGCAUGAUGCGGAAUACUUGCACCACCCGGCGCACAUCCUGGAACUGGUCCGUUGCAUUCUUAUUCGUUUCCAAUAAAAAUAACG (..(((((((((........(((.((((((((((.....))).......(((.((....)).))))))))))))))))))))))..)............................. ( -40.80, z-score = -2.84, R) >droGri2.scaffold_15110 19503593 116 - 24565398 GUGACGGGCCAGCUUAAGGAUUCGCAGGAUGCGCAUGAUGCGGAAUACUUGCACCACCCGGCGCACAUCCUGGAACUGGUCCGUUGCAUUCUUAUUCGUUUCCAAUAAAAAUAACG (..(((((((((........(((.((((((((((.....))).......(((.((....)).))))))))))))))))))))))..)............................. ( -40.80, z-score = -2.84, R) >anoGam1.chr2L 25639251 116 + 48795086 AUGGCGGGCCAGCUUGAGGAUUCGCAGGAUGCGCAUGAUGCGGAAGACUUGCACCACCCGGCGCACAUCCUGGAACUGGUCCGUCGCGUUCUUGUUGGUCUCCAACAGGAACAGGG ..((((((((((.((.((((((((((..((....))..)))))).....(((.((....)).)))..)))).)).))))))))))..(((((((((((...))))))))))).... ( -54.60, z-score = -3.41, R) >apiMel3.GroupUn 83739118 116 - 399230636 GUGGCGGGCCAGCUUCAGGAUCCUCAGGAUCCGCAUUAUACGAAAGAUUUGCACUACCCUGCGCACGUCCUGGAACUGGUCGGUCGCAUUCUUGUUCGUCUCGACCAGGAACAGAG (((((.((((((.((((((((.(.((((....(((..((.(....))).))).....)))).)...)))))))).)))))).))))).((((((.(((...))).))))))..... ( -45.20, z-score = -2.80, R) >triCas2.ChLG8 12910648 103 - 15773733 AUGUCGAGUCAGUUUAAACAGUCGCAUGAUUCGAAUUAUUGAAAAGAAUUCCAUUAUGUCG---GCGUUUUCUAAGUGGGCCG-CGUACCGACGGAGGAUCAAAUCG--------- ...((((((((((..........)).))))))))...........((..(((....(((((---((((..(((....)))..)-))..))))))..)))))......--------- ( -22.00, z-score = 0.69, R) >consensus GUGACGGGCCAGCUUAAGGAUUCGCAGGAUGCGCAUGAUGCGAAAGACUUGCACCACCCGGCGCACAUCCUGGAACUGGUCCGUUGCAUUCUUAUUCGUUUCCAAUAGAAAUAACG (((((((((((..((.((((((((((..((....))..)))))).....(((..........)))..)))).))..)))))))))))............................. (-22.92 = -22.95 + 0.03)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:59:09 2011