
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 2,880,788 – 2,880,950 |
| Length | 162 |
| Max. P | 0.944741 |

| Location | 2,880,788 – 2,880,950 |
|---|---|
| Length | 162 |
| Sequences | 8 |
| Columns | 169 |
| Reading direction | forward |
| Mean pairwise identity | 74.68 |
| Shannon entropy | 0.53554 |
| G+C content | 0.41146 |
| Mean single sequence MFE | -39.13 |
| Consensus MFE | -20.94 |
| Energy contribution | -22.46 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.855050 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
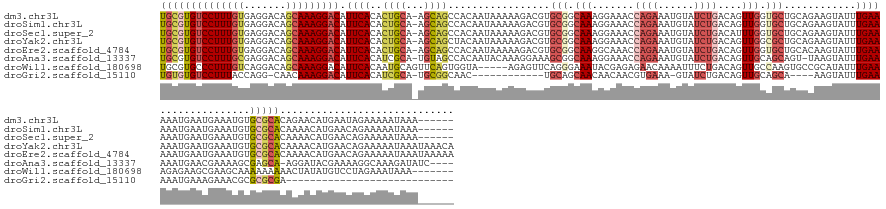
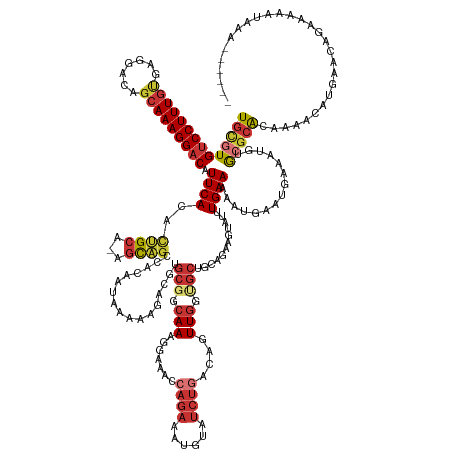
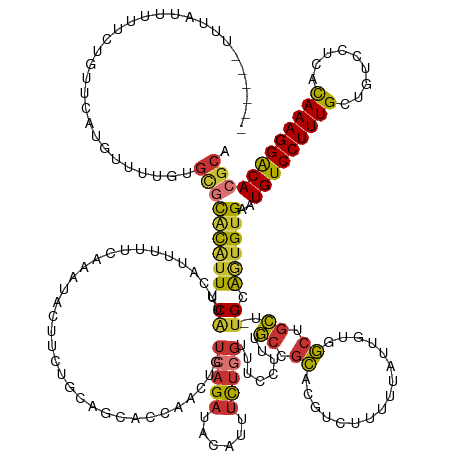
>dm3.chr3L 2880788 162 + 24543557 UGCGUGUCCUUUGUGAGGACAGCAAAGGACAUUCACACUGCA-AGCAGCCACAAUAAAAAGACGUGCGGCAAAGGAAACCAGAAAUGUAUCUGACAGUUGGUGCUGCAGAAGUAUUUGAAAAAUGAAUGAAAUGUGCGCACAGAACAUGAAUAGAAAAAUAAA------ ((((((((((((((.......)))))))))(((((..(((((-.((.(((.((...........)).))).......(((((...(((.....))).))))))))))))......((....))))))).......))))).......................------ ( -41.40, z-score = -1.69, R) >droSim1.chr3L 2433756 162 + 22553184 UGCGUGUCCUUUGUGAGGACAGCAAAGGACAUUCACACUGCA-AGCAGCCACAAUAAAAAGACGUGCGGCAAAGGAAACCAGAAAUGUAUCUGACAGUUGGUGCUGCAGAAGUAUUUGAAAAAUGAAUGAAAUGUGCGCACAAAACAUGAACAGAAAAAUAAA------ ((((((((((((((.......)))))))))(((((..(((((-.((.(((.((...........)).))).......(((((...(((.....))).))))))))))))......((....))))))).......))))).......................------ ( -41.40, z-score = -1.60, R) >droSec1.super_2 2900928 162 + 7591821 UGCGUGUCCUUUGUGAGGACAGCAAAGGACAUUCACACUGCA-AGCAGCCACAAUAAAAAGACGUGCGGCAAAGGAAACCAGAAAUGUAUCUGACAUUUGGUGCUGCAGAAGUAUUUGAAAAAUGAAUGAAAUGUGCGCACAAAACAUGAACAGAAAAAUAAA------ ((((((((((((((.......)))))))))(((((..(((((-.((.(((.((...........)).))).......((((..(((((.....))))))))))))))))......((....))))))).......))))).......................------ ( -40.90, z-score = -1.56, R) >droYak2.chr3L 15147716 168 - 24197627 UGCGUGUCCUUUGUGAGGACAGCAAAGGACAUUCACACUGCA-AGCAGCUACAAUAAAAAGACGUGCGGCAAAGGAAACCAGAAAUGUAUCUGACAGUUGGCGCUGCAGAAGUAUUUGAAAAAUGAAUGAAAUGUGCGCACAAAACAUGAACAGAAAAAUAAAUAAACA ((.(((((((((((.......))))))))))).))..(((..-..)))...............((((((((..(....)...(((((..((((.((((....))))))))..)))))...............))).)))))............................ ( -39.50, z-score = -1.06, R) >droEre2.scaffold_4784 2907565 168 + 25762168 UGCGUGUCCUUUGUGAGGACAGCAAAGGACAUUCACACUGCA-AGCAGCCACAAUAAAAAGACGUGCGGCAAGGCAAACCAGAAAUGUAUCUGACAGUUGGUGCUGCACAAGUAUUUGAAAAAUGAAUGAAAUGUGCGCACAAAACAUGAACAGAAAAAUAAAUAAAAA ((((((((((((((.......)))))))))(((((..(((..-..)))...............((((((((.(((....((((......))))...)))..))))))))..............))))).......)))))............................. ( -42.40, z-score = -1.48, R) >droAna3.scaffold_13337 14086429 162 - 23293914 UGCGUGUCCUUUGCGAGGACAGCAAAGGACAUUCACAUCGCA-UGUAGCCACAAUACAAAGGAAAGCGGCAAAGGAAACCAGAAAUGUAUCUGACAGUUGCAGCAGU-UAAGUAUUUGAAAAAUGAACGAAAAGCGAGCA-AGGAUACGAAAAGGCAAAGAUAUC---- ((((((((((((((.......)))))))))).......))))-....(((...............((.((((.(....)((((......))))....)))).))...-...((((((......((..((.....))..))-.)))))).....))).........---- ( -38.81, z-score = -1.85, R) >droWil1.scaffold_180698 351708 157 - 11422946 UGCGUGCCCUUUGUCAGGACAGCAAAGGACAUUCACAAUGCAGUUCAGUGGUA-----AGAGUUCAGGGAAAUACGAGAGAACAAAAUUUCUGACAGUUGCCAAGUGCCGCAUAUUUGAAAGAGAAGCGAAGCAAAAAAAAACUAUAUGUCCUAGAAAUAAA------- (((.((.(((.....))).))))).((((((((((..((((.((....(((((-----(..((.(((..(....(....).......)..)))))..))))))...)).))))...))))......((...))..............)))))).........------- ( -33.20, z-score = -0.22, R) >droGri2.scaffold_15110 19448938 122 + 24565398 UGUGUGUCCUUUACCAGG-CAACAAAGGACAUUCACAUCGCA-UGCGGCAAC------------UGCAGCAACAACAACGUGAAA-GUAUCUGACAGUUGCAGCA----AAGUAUUUGAAAAAUGAAAGAAACGCGCGCGA---------------------------- ((((((((((((....(.-...)))))))))..))))((((.-(((((((((------------(((((..((............-))..))).)))))))..((----(.....)))..............)))).))))---------------------------- ( -35.40, z-score = -1.49, R) >consensus UGCGUGUCCUUUGUGAGGACAGCAAAGGACAUUCACACUGCA_AGCAGCCACAAUAAAAAGACGUGCGGCAAAGGAAACCAGAAAUGUAUCUGACAGUUGGUGCUGCAGAAGUAUUUGAAAAAUGAAUGAAAUGUGCGCACAAAACAUGAACAGAAAAAUAAA______ ((((((((((((((.......))))))))).((((..((((...)))).................(((.(((.......((((......))))....))).)))............))))...............)))))............................. (-20.94 = -22.46 + 1.52)
| Location | 2,880,788 – 2,880,950 |
|---|---|
| Length | 162 |
| Sequences | 8 |
| Columns | 169 |
| Reading direction | reverse |
| Mean pairwise identity | 74.68 |
| Shannon entropy | 0.53554 |
| G+C content | 0.41146 |
| Mean single sequence MFE | -38.35 |
| Consensus MFE | -19.73 |
| Energy contribution | -19.78 |
| Covariance contribution | 0.05 |
| Combinations/Pair | 1.45 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.944741 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
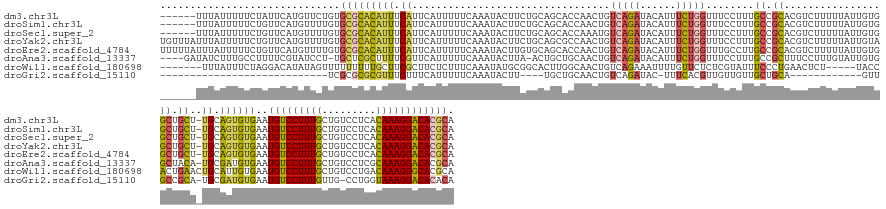
>dm3.chr3L 2880788 162 - 24543557 ------UUUAUUUUUCUAUUCAUGUUCUGUGCGCACAUUUCAUUCAUUUUUCAAAUACUUCUGCAGCACCAACUGUCAGAUACAUUUCUGGUUUCCUUUGCCGCACGUCUUUUUAUUGUGGCUGCU-UGCAGUGUGAAUGUCCUUUGCUGUCCUCACAAAGGACACGCA ------.......................((((......................(((..((((((((.......(((((......)))))........((((((...........))))))))).-))))).)))..(((((((((.((....))))))))))))))) ( -39.80, z-score = -2.41, R) >droSim1.chr3L 2433756 162 - 22553184 ------UUUAUUUUUCUGUUCAUGUUUUGUGCGCACAUUUCAUUCAUUUUUCAAAUACUUCUGCAGCACCAACUGUCAGAUACAUUUCUGGUUUCCUUUGCCGCACGUCUUUUUAUUGUGGCUGCU-UGCAGUGUGAAUGUCCUUUGCUGUCCUCACAAAGGACACGCA ------.......................((((......................(((..((((((((.......(((((......)))))........((((((...........))))))))).-))))).)))..(((((((((.((....))))))))))))))) ( -39.80, z-score = -2.03, R) >droSec1.super_2 2900928 162 - 7591821 ------UUUAUUUUUCUGUUCAUGUUUUGUGCGCACAUUUCAUUCAUUUUUCAAAUACUUCUGCAGCACCAAAUGUCAGAUACAUUUCUGGUUUCCUUUGCCGCACGUCUUUUUAUUGUGGCUGCU-UGCAGUGUGAAUGUCCUUUGCUGUCCUCACAAAGGACACGCA ------.......................((((......................(((..((((((((.......(((((......)))))........((((((...........))))))))).-))))).)))..(((((((((.((....))))))))))))))) ( -39.80, z-score = -1.98, R) >droYak2.chr3L 15147716 168 + 24197627 UGUUUAUUUAUUUUUCUGUUCAUGUUUUGUGCGCACAUUUCAUUCAUUUUUCAAAUACUUCUGCAGCGCCAACUGUCAGAUACAUUUCUGGUUUCCUUUGCCGCACGUCUUUUUAUUGUAGCUGCU-UGCAGUGUGAAUGUCCUUUGCUGUCCUCACAAAGGACACGCA .................((((((.....((((...........................(((((((......))).)))).........(((.......)))))))..............((((..-..))))))))))((((((((.((....))))))))))..... ( -36.00, z-score = -0.77, R) >droEre2.scaffold_4784 2907565 168 - 25762168 UUUUUAUUUAUUUUUCUGUUCAUGUUUUGUGCGCACAUUUCAUUCAUUUUUCAAAUACUUGUGCAGCACCAACUGUCAGAUACAUUUCUGGUUUGCCUUGCCGCACGUCUUUUUAUUGUGGCUGCU-UGCAGUGUGAAUGUCCUUUGCUGUCCUCACAAAGGACACGCA .................((((((.....(((((((((......................))))).))))..(((((((((......))))....((...((((((...........)))))).)).-.)))))))))))((((((((.((....))))))))))..... ( -44.25, z-score = -2.44, R) >droAna3.scaffold_13337 14086429 162 + 23293914 ----GAUAUCUUUGCCUUUUCGUAUCCU-UGCUCGCUUUUCGUUCAUUUUUCAAAUACUUA-ACUGCUGCAACUGUCAGAUACAUUUCUGGUUUCCUUUGCCGCUUUCCUUUGUAUUGUGGCUACA-UGCGAUGUGAAUGUCCUUUGCUGUCCUCGCAAAGGACACGCA ----(((((............)))))..-(((((((...((((..........(((((...-...((.((((...(((((......)))))......)))).))........))))).........-.)))).)))).((((((((((.......)))))))))).))) ( -37.79, z-score = -2.36, R) >droWil1.scaffold_180698 351708 157 + 11422946 -------UUUAUUUCUAGGACAUAUAGUUUUUUUUUGCUUCGCUUCUCUUUCAAAUAUGCGGCACUUGGCAACUGUCAGAAAUUUUGUUCUCUCGUAUUUCCCUGAACUCU-----UACCACUGAACUGCAUUGUGAAUGUCCUUUGCUGUCCUGACAAAGGGCACGCA -------.........(((((((................((((...............))))(((...(((....((((.......((((..............))))...-----.....))))..)))...))).))))))).(((((((((.....)))))).))) ( -34.36, z-score = -1.25, R) >droGri2.scaffold_15110 19448938 122 - 24565398 ----------------------------UCGCGCGCGUUUCUUUCAUUUUUCAAAUACUU----UGCUGCAACUGUCAGAUAC-UUUCACGUUGUUGUUGCUGCA------------GUUGCCGCA-UGCGAUGUGAAUGUCCUUUGUUG-CCUGGUAAAGGACACACA ----------------------------(((((((((.......)...............----(((.(((((((.(((..((-............))..)))))------------))))).)))-.))).))))).(((((((((...-.....))))))))).... ( -35.00, z-score = -1.11, R) >consensus ______UUUAUUUUUCUGUUCAUGUUUUGUGCGCACAUUUCAUUCAUUUUUCAAAUACUUCUGCAGCACCAACUGUCAGAUACAUUUCUGGUUUCCUUUGCCGCACGUCUUUUUAUUGUGGCUGCU_UGCAGUGUGAAUGUCCUUUGCUGUCCUCACAAAGGACACGCA ..............................(((((((((.((.................................(((((......)))))........((.((................)).))..)).))))))..(((((((((.((....)))))))))))))). (-19.73 = -19.78 + 0.05)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:58:56 2011