| Sequence ID | dm3.chr3L |
|---|---|
| Location | 2,875,260 – 2,875,365 |
| Length | 105 |
| Max. P | 0.765842 |

| Location | 2,875,260 – 2,875,365 |
|---|---|
| Length | 105 |
| Sequences | 7 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 73.97 |
| Shannon entropy | 0.47438 |
| G+C content | 0.48166 |
| Mean single sequence MFE | -29.97 |
| Consensus MFE | -13.62 |
| Energy contribution | -14.00 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.62 |
| SVM RNA-class probability | 0.765842 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 2875260 105 + 24543557 UUAGAAUCUA-GGUGACUGCGAGGGCUGUUUUUUGGCAGUA----UAUACAUAUGUGUAUUAUGUGGAGGGGCAGGGAGCCUCACAGAACGAGGAACUAGGCUACACACA ((((..(((.-.((..(((.((((.(((((((((.(((..(----(((((....))))))..))).)))))))))....)))).))).)).)))..)))).......... ( -29.50, z-score = -1.40, R) >droSim1.chr3L 2428181 109 + 22553184 UUAGAAUCUA-GGUGACUGCGAGGGCUGUUUUUUGGCAGUAUAUACAUACAUAUGUGUAUUAUGUGGAGGGGCAGGGAGCCUCACAGGACGAGGAACUAGGCUACACACA .(((...(((-(.((.(((.((((.(((((((((.(((..((((((((....))))))))..))).)))))))))....)))).)))..)).....)))).)))...... ( -32.20, z-score = -2.07, R) >droSec1.super_2 2895374 109 + 7591821 UUAGAAUCUA-GGUGACUGCGAGGGCUGUUUUUUGGCAGUAUAUACAUACAUAUGUGUAUUGUGUGGAGGGGCAGGGAGCCUCACAGGACGAGGAACUAGGCUACACACA ......((((-(....))).))..((((((....))))))((((((((....))))))))((((((....(((..((..((((.......))))..))..))).)))))) ( -34.10, z-score = -2.25, R) >droYak2.chr3L 15142024 109 - 24197627 AUAGAAUCUA-GGUGACUGCGAGGGCUGUUUUUUGGCAGUACAUAUACAUAUGUGUGUUUUAUGUAGAGGGGCAGGGAGCCUCGCAGGACGAGGAACUAGGCUACACACA .(((...(((-(.((.((((((((.(((((((((.(((..(((((((....)))))))....))).)))))))))....))))))))..)).....)))).)))...... ( -34.00, z-score = -2.16, R) >droEre2.scaffold_4784 2901981 105 + 25762168 AUAGAAUCUA-GGUGACUGCGAGGGCUGUUUUUUGGCAGUA--UAUACAUACGUGUAUU--AUGUGGAGGGGCAGGGAGCCUCGCAGGACGAGGAACUAGGCUACACACA .(((...(((-(.((.((((((((.(((((((((.(((..(--(((((....)))))).--.))).)))))))))....))))))))..)).....)))).)))...... ( -35.50, z-score = -3.03, R) >dp4.chrXR_group8 3714103 83 + 9212921 ---GAGUCGAGGGCGAGGGCUG----UUUUUUUU-----------CGGGUAUCUGUAUC-----UAAAGGAACAGAGAG----AGCGGAAGAAGAACUAGGCUACACACA ---.((((...(((....)))(----((((((((-----------(.(.(.(((((.((-----....)).)))))...----).).))))))))))..))))....... ( -23.60, z-score = -1.90, R) >droPer1.super_23 1629131 87 + 1662726 ---GAGUCGACGGCGAGGGCAGGGGCUUUUUUUU-----------CGGGUAUCUGUAUC-----UAAAGGAACAGAGAG----AGCGGAAGAAGAACUAGGCUACACACA ---...(((....))).(((...((.((((((((-----------(.(.(.(((((.((-----....)).)))))...----).).)))))))))))..)))....... ( -20.90, z-score = -0.61, R) >consensus UUAGAAUCUA_GGUGACUGCGAGGGCUGUUUUUUGGCAGUA__UACAUAUAUAUGUGUAUUAUGUGGAGGGGCAGGGAGCCUCACAGGACGAGGAACUAGGCUACACACA ............(((...((.....(((((((((.(((.......(((....))).......))).)))))))))((..((((.......))))..))..))..)))... (-13.62 = -14.00 + 0.37)

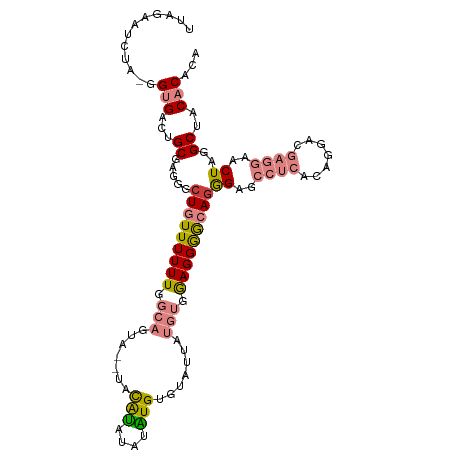

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:58:54 2011