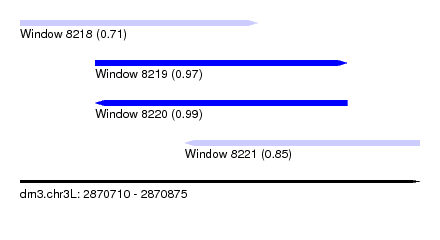
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 2,870,710 – 2,870,875 |
| Length | 165 |
| Max. P | 0.994040 |

| Location | 2,870,710 – 2,870,808 |
|---|---|
| Length | 98 |
| Sequences | 3 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 82.05 |
| Shannon entropy | 0.24030 |
| G+C content | 0.60017 |
| Mean single sequence MFE | -42.70 |
| Consensus MFE | -32.61 |
| Energy contribution | -32.73 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.714520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 2870710 98 + 24543557 UUUGCAGGGAACUCAUGGCGGUGGG---------UCCCACGGGUCCUGCUACUGAAUUGGGUCCUACGGAGCCAGGAGGUAUGGAACCUGUUGCAGCUGCUACUGCA .(((((((((.(((((....)))))---------))))((((((((...((((...((((.(((...))).))))..)))).)).)))))))))))..((....)). ( -38.30, z-score = -0.72, R) >droYak2.chr3L 15137306 107 - 24197627 UUUGCAGCGAGCUCAUGGCAGUGGGACCCACAGAUCCCACGGAUCCCGUUACUGAGUUUGGUCCCACGGAGCCAGGAGGUAGGACACCUGCAGCAGCUGCGACCGCU .(((((((...(((.((((.((((((((.((.(((((...))))).((....)).))..))))))))...)))).)))(((((...)))))....)))))))..... ( -45.50, z-score = -2.40, R) >droSec1.super_2 2890877 98 + 7591821 UUUGCAGGGAGCUCAUGGCGGUGGG---------UCCCACGGGUCCCACUACUGAAUUGGGACCUACGGAGCCAGGAGGUAUGGAACCUGCAGCAGCUGCUACUGCU ..((((((.(.(((.((((.(((((---------((((((((.........)))...))))))))))...)))).))).)......))))))((((......)))). ( -44.30, z-score = -2.19, R) >consensus UUUGCAGGGAGCUCAUGGCGGUGGG_________UCCCACGGGUCCCGCUACUGAAUUGGGUCCUACGGAGCCAGGAGGUAUGGAACCUGCAGCAGCUGCUACUGCU ...(((((.(.(((.((((.(((((..........)))))(((.((((.........)))).))).....)))).))).)......)))))(((((......))))) (-32.61 = -32.73 + 0.12)

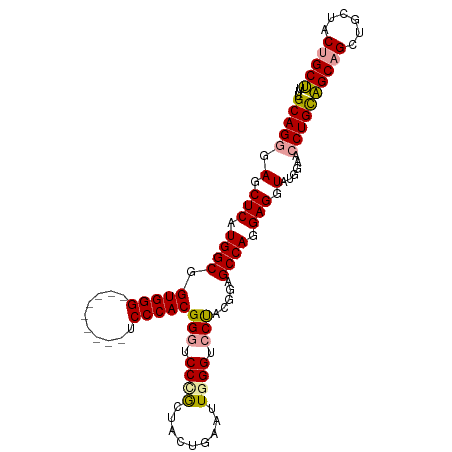
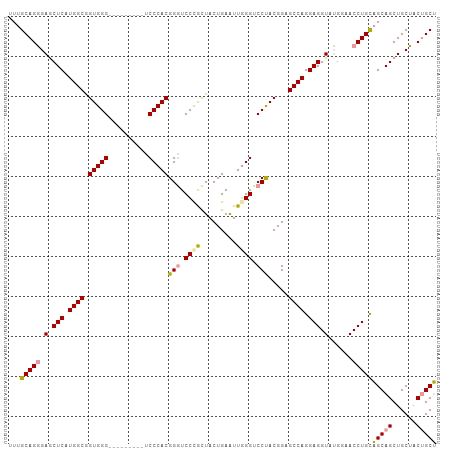
| Location | 2,870,741 – 2,870,845 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 64.11 |
| Shannon entropy | 0.58686 |
| G+C content | 0.62946 |
| Mean single sequence MFE | -44.80 |
| Consensus MFE | -24.25 |
| Energy contribution | -25.38 |
| Covariance contribution | 1.12 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.85 |
| SVM RNA-class probability | 0.971540 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 2870741 104 + 24543557 GGGUCCUGCUACUGAAUUGGGUCCUACGGAGCCAGGAGGUAUGGAACCUGUUGCAGCUGCUA-CUGCAGCUGCCGAUCCUGGUGGCACUCCUGCUCCUAUGGGCA ((..((............))..))...(((((.(((((((.....(((.(((((((((((..-..))))))).))))...))).)).))))))))))........ ( -45.00, z-score = -1.48, R) >anoGam1.chr2L 5454804 91 - 48795086 ------------GGCAGCGGUGGCAGCGGAGUCGGUGUCAGCUGCGUUAGCUGUGUCAGCGAGCCGCCCGUACCGGGUGAGGCGGAGCUC--GGUGAGAUCGGUA ------------.(((((..(((((.((....)).))))))))))....(((((.(((.(((((((((((...)))))).......))))--).))).).)))). ( -40.31, z-score = -0.86, R) >droYak2.chr3L 15137346 104 - 24197627 GGAUCCCGUUACUGAGUUUGGUCCCACGGAGCCAGGAGGUAGGACACCUGCAGCAGCUGCGA-CCGCUGCUGCCGAUCCAGGUGGCACUCCUGCUCCUAUGGGCA ....................((((...(((((.(((((....(.(((((((((((((.....-..))))))))......))))).).))))))))))...)))). ( -43.80, z-score = -0.79, R) >droSec1.super_2 2890908 104 + 7591821 GGGUCCCACUACUGAAUUGGGACCUACGGAGCCAGGAGGUAUGGAACCUGCAGCAGCUGCUA-CUGCUGCUGCCGAUCCUGGUGGCACUCCUGCUCCUAUGGGCA ((((((((.........))))))))..(((((.(((((((..(((.(..((((((((.....-..)))))))).).))).....)).))))))))))........ ( -50.10, z-score = -3.29, R) >consensus GGGUCCCGCUACUGAAUUGGGUCCUACGGAGCCAGGAGGUAUGGAACCUGCAGCAGCUGCGA_CCGCUGCUGCCGAUCCAGGUGGCACUCCUGCUCCUAUGGGCA ....................((((...(((((.(((((...((.((((..(.(((((.((.....)).))))).).....)))).))))))))))))...)))). (-24.25 = -25.38 + 1.12)

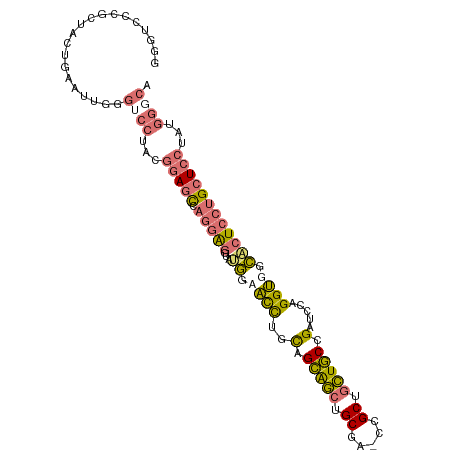
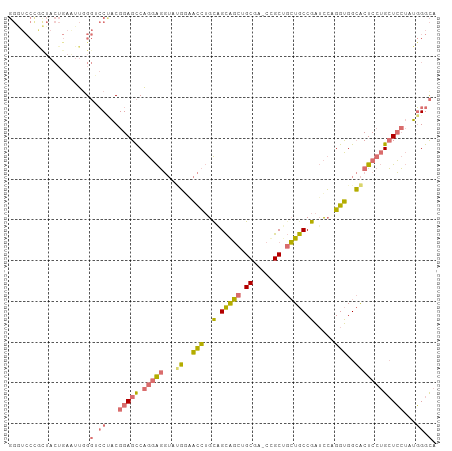
| Location | 2,870,741 – 2,870,845 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 64.11 |
| Shannon entropy | 0.58686 |
| G+C content | 0.62946 |
| Mean single sequence MFE | -41.05 |
| Consensus MFE | -22.11 |
| Energy contribution | -26.30 |
| Covariance contribution | 4.19 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.66 |
| SVM RNA-class probability | 0.994040 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 2870741 104 - 24543557 UGCCCAUAGGAGCAGGAGUGCCACCAGGAUCGGCAGCUGCAG-UAGCAGCUGCAACAGGUUCCAUACCUCCUGGCUCCGUAGGACCCAAUUCAGUAGCAGGACCC (((((...(((((.((....))..(((((...((((((((..-..))))))))....(((.....)))))))))))))...))((........)).)))...... ( -38.80, z-score = -1.50, R) >anoGam1.chr2L 5454804 91 + 48795086 UACCGAUCUCACC--GAGCUCCGCCUCACCCGGUACGGGCGGCUCGCUGACACAGCUAACGCAGCUGACACCGACUCCGCUGCCACCGCUGCC------------ ........(((.(--((((..(((((..........)))))))))).)))..((((....(((((.((.......)).)))))....))))..------------ ( -29.30, z-score = -1.31, R) >droYak2.chr3L 15137346 104 + 24197627 UGCCCAUAGGAGCAGGAGUGCCACCUGGAUCGGCAGCAGCGG-UCGCAGCUGCUGCAGGUGUCCUACCUCCUGGCUCCGUGGGACCAAACUCAGUAACGGGAUCC ..(((((.((((((((((.(.((((((...(((((((.((..-..)).))))))))))))).)....))))).))))))))))......(((......))).... ( -47.80, z-score = -1.89, R) >droSec1.super_2 2890908 104 - 7591821 UGCCCAUAGGAGCAGGAGUGCCACCAGGAUCGGCAGCAGCAG-UAGCAGCUGCUGCAGGUUCCAUACCUCCUGGCUCCGUAGGUCCCAAUUCAGUAGUGGGACCC ........((((((((((.(......(((((.((((((((..-.....)))))))).)))))....)))))).)))))...(((((((.........))))))). ( -48.30, z-score = -3.54, R) >consensus UGCCCAUAGGAGCAGGAGUGCCACCAGGAUCGGCAGCAGCAG_UAGCAGCUGCAGCAGGUGCCAUACCUCCUGGCUCCGUAGGACCCAAUUCAGUAGCGGGACCC ..(((((.((((((((((((.((((.....((((((((((.....))))))))))..)))).)))...))))).)))))))))...................... (-22.11 = -26.30 + 4.19)


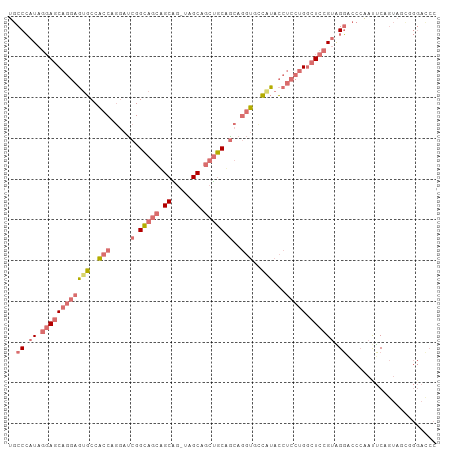
| Location | 2,870,778 – 2,870,875 |
|---|---|
| Length | 97 |
| Sequences | 3 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 93.13 |
| Shannon entropy | 0.09467 |
| G+C content | 0.58076 |
| Mean single sequence MFE | -37.97 |
| Consensus MFE | -33.65 |
| Energy contribution | -33.77 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.91 |
| SVM RNA-class probability | 0.849980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 2870778 97 - 24543557 AAUUACUACCGGAUGGAACACCAAUGCCUGUGCCCAUAGGAGCAGGAGUGCCACCAGGAUCGGCAGCUGCAGUAGCAGCUGCAACAGGUUCCAUACC ..........(((((((((..((...(((((.((....)).)))))..))((....))....((((((((....)))))))).....))))))).)) ( -37.20, z-score = -2.45, R) >droYak2.chr3L 15137383 97 + 24197627 AAUUACUACCGGAUGGAACACCCAUGCCUGUGCCCAUAGGAGCAGGAGUGCCACCUGGAUCGGCAGCAGCGGUCGCAGCUGCUGCAGGUGUCCUACC ..........((..(((.(((((((.(((((.((....)).))))).)))((....))....((((((((.......)))))))).)))))))..)) ( -38.00, z-score = -0.91, R) >droSec1.super_2 2890945 97 - 7591821 AAUUACUACCGGAUGGAACACCCAUGCCUGUGCCCAUAGGAGCAGGAGUGCCACCAGGAUCGGCAGCAGCAGUAGCAGCUGCUGCAGGUUCCAUACC ..........(((((((((.......(((((.((....)).)))))..((((.........))))((((((((....))))))))..))))))).)) ( -38.70, z-score = -2.33, R) >consensus AAUUACUACCGGAUGGAACACCCAUGCCUGUGCCCAUAGGAGCAGGAGUGCCACCAGGAUCGGCAGCAGCAGUAGCAGCUGCUGCAGGUUCCAUACC ..........(((((((((.......(((((.((....)).)))))..((((.........))))((((((((....))))))))..))))))).)) (-33.65 = -33.77 + 0.12)

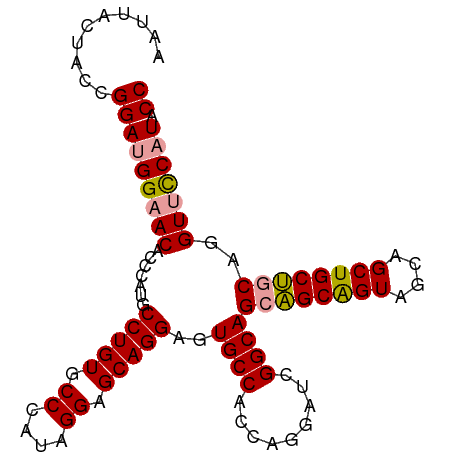
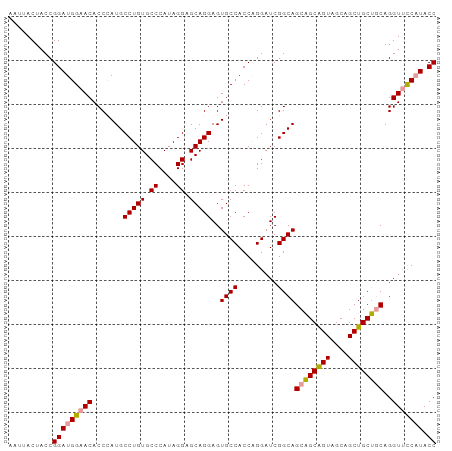
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:58:51 2011