| Sequence ID | dm3.chr3L |
|---|---|
| Location | 2,870,251 – 2,870,348 |
| Length | 97 |
| Max. P | 0.800377 |

| Location | 2,870,251 – 2,870,348 |
|---|---|
| Length | 97 |
| Sequences | 9 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 74.97 |
| Shannon entropy | 0.50454 |
| G+C content | 0.62044 |
| Mean single sequence MFE | -33.57 |
| Consensus MFE | -17.28 |
| Energy contribution | -17.77 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.73 |
| SVM RNA-class probability | 0.800377 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
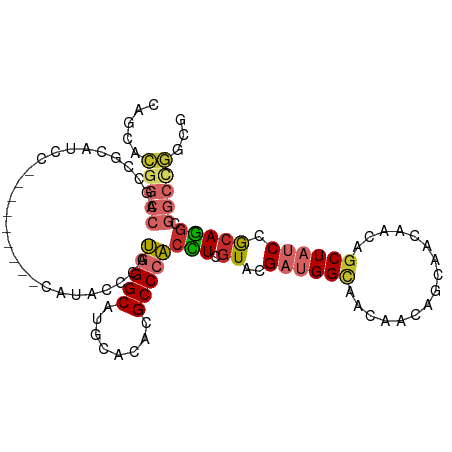
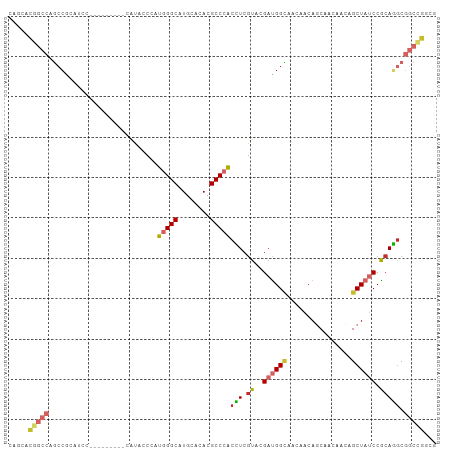
>dm3.chr3L 2870251 97 - 24543557 CAGCACGGCCAGCCGCAUCC---------CAUACCAAUGGGCAUGCACACGCCCACCUCGUAUGAUGGCAACAACAGCAACAACAGCUAUCCGCAGGCGGCUGGCG .......(((((((((((((---------(((....))))).))))....(((......((.((.((....)).)))).......((.....)).)))))))))). ( -38.50, z-score = -2.91, R) >droSim1.chr3L 2423255 97 - 22553184 CAGCACGGCCAGCCGCAUCC---------CAUACCCAUGGGCAUGCACACGCCCACCUCGUACGAUGGCAACAACAGCAACAACAGCUAUCCGCAGGCGGCUGGCG .......(((((((((((((---------(((....))))).))))....(((..........(.((....)).)(((.......))).......)))))))))). ( -37.40, z-score = -2.64, R) >droSec1.super_2 2890418 97 - 7591821 CAGCACGGCCAGCCGCAUCC---------CAUACCCAUGGGCAUGCACACGCCCACCUCGUACGAUGGCAACAACAGCAACAACAGCUAUCCGCAGGCGGCUGGCG .......(((((((((((((---------(((....))))).))))....(((..........(.((....)).)(((.......))).......)))))))))). ( -37.40, z-score = -2.64, R) >droYak2.chr3L 15136847 97 + 24197627 CAGCACGGCCAGCCGCAUCC---------CAUACCCAUGGGCAUGCACACGCCCACCUCGUACGAUGGCAACAACAGCAACAACAGCUAUCCACAGGCGGCAGGCG .......(((.(((((((((---------(((....))))).))))....(((..........(.((....)).)(((.......))).......)))))).))). ( -32.10, z-score = -1.94, R) >droEre2.scaffold_4784 2896829 97 - 25762168 CAGCACGGCCAGCCGCAUCC---------CAUACCCAUGGGCAUGCACACGCCCACCUCGUACGAUGGCAACAACAGCAACAACAGCUAUCCGCAGGCGGCUGGAG ........((((((((((((---------(((....))))).))))....(((..........(.((....)).)(((.......))).......))))))))).. ( -33.60, z-score = -1.78, R) >droAna3.scaffold_13337 14075680 91 + 23293914 CAGCACCACCAGCCGCACCC---------CAUUCCCAUGGGCAUGCACACGCCGACCUCGUUCGACGGCA---ACAGCAACACCAGCUACCCGCAGACGGCCA--- ...........(((((..((---------(((....)))))..(((....((((...((....)))))).---..(((.......)))....)))).))))..--- ( -23.30, z-score = -1.08, R) >droWil1.scaffold_180698 340700 88 + 11422946 CAGAAUGUAGGGCA---------------CGUUCCACAUGGCUUGCAUACGCCCUCAUCAUACGAUGGUA---ACAGCCAGAAUAGCUAUCCACAUGCAACCACCA ..((((((.....)---------------)))))....(((.((((((......(((((....)))))..---..(((.......)))......)))))))))... ( -17.90, z-score = 0.30, R) >droVir3.scaffold_13049 15006284 100 - 25233164 CAACAUGGGCACACACAUCCCGGCCUGGGCGUGGCUGUGGGCAUGCAUACGCCCACAUCGUACGAUGGCA---ACAACAGCAACAGCUAUCCACAUGCCGCCA--- ......(((.........)))(((...(((((((.(((((((........))))))).)....((((((.---............))))))..))))))))).--- ( -35.42, z-score = -0.52, R) >droMoj3.scaffold_6680 2883658 100 + 24764193 CUGCAUGGCCAUCCUGGCCUGGGUGUGGGCGUGGCUGUGGGCAUGCACACGCCCACGUCGUACGACGGGA---ACAACAGCAACAGCUUCCCGCACGCGGCCA--- ((((..((((.....))))...(((((((...((((((((((........)))).((((....))))...---.........)))))).)))))))))))...--- ( -46.50, z-score = -0.74, R) >consensus CAGCACGGCCAGCCGCAUCC_________CAUACCCAUGGGCAUGCACACGCCCACCUCGUACGAUGGCAACAACAGCAACAACAGCUAUCCGCAGGCGGCCGGCG .....(((((...........................(((((........)))))(((.((..((((((................)))))).))))).)))))... (-17.28 = -17.77 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:58:49 2011