| Sequence ID | dm3.chr3L |
|---|---|
| Location | 2,856,707 – 2,856,811 |
| Length | 104 |
| Max. P | 0.961899 |

| Location | 2,856,707 – 2,856,811 |
|---|---|
| Length | 104 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 85.67 |
| Shannon entropy | 0.29415 |
| G+C content | 0.31095 |
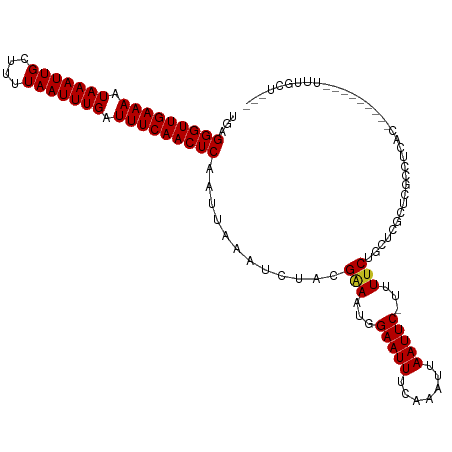
| Mean single sequence MFE | -21.03 |
| Consensus MFE | -16.00 |
| Energy contribution | -16.02 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.961899 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
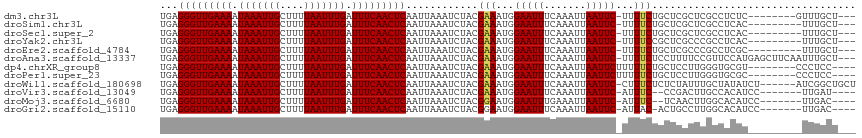
>dm3.chr3L 2856707 104 - 24543557 UGAGGGUUGAAAAUAAAUUGCUUUUAAUUUGAUUUCAACUCAAUUAAAUCUACGAAAUGGAAUUUCAAAUUAAUUC-UUUUCUGCUCGCUCGCCUCUC--------GUUUGCU--- .(((((((((((.(((((((....))))))).)))))))..............((((.((((((.......)))))-)))))..........))))..--------.......--- ( -20.00, z-score = -1.94, R) >droSim1.chr3L 2409817 103 - 22553184 UGAGGGUUGAAAAUAAAUUGCUUUUAAUUUGAUUUCAACUCAAUUAAAUCUACGAAAUGGAAUUUCAAAUUAAUUC-UUUUCUGCUCGCUCGCCUCAC---------UUUGCU--- ((((((((((((.(((((((....))))))).)))))))..............((((.((((((.......)))))-)))))..........))))).---------......--- ( -20.90, z-score = -2.66, R) >droSec1.super_2 2877003 103 - 7591821 UGAGGGUUGAAAAUAAAUUGCUUUUAAUUUGAUUUCAACUCAAUUAAAUCUACGAAAUGGAAUUUCAAAUUAAUUC-UUUUCUGCUCGCUCGCCUCAC---------UUUGCU--- ((((((((((((.(((((((....))))))).)))))))..............((((.((((((.......)))))-)))))..........))))).---------......--- ( -20.90, z-score = -2.66, R) >droYak2.chr3L 15122545 103 + 24197627 UGAGGGUUGAAAAUAAAUUGCUUUUAAUUUGAUUUCAACUCAAUUAAAUCUACGAAAUGGAAUUUCAAAUUAAUUC-UUUUCCGCUCGCCCGCCUCAC---------UUUGCU--- ((((((((((((.(((((((....))))))).)))))))..............((((.((((((.......)))))-)))))..........))))).---------......--- ( -20.50, z-score = -2.19, R) >droEre2.scaffold_4784 2883117 103 - 25762168 UGAGGGUUGAAAAUAAAUUGCUUUUAAUUUGAUUUCAACUCAAUUAAAUCUACGAAAUGGAAUUUCAAAUUAAUUC-UUUUCUGCUCGCCCGCCUCGC---------UUUGCU--- .(((((((((((.(((((((....))))))).)))))))..............((((.((((((.......)))))-)))))..........))))..---------......--- ( -20.50, z-score = -1.89, R) >droAna3.scaffold_13337 14060967 112 + 23293914 UGAGGGUUGAAAAUAAAUUGCUUUUAAUUUGAUUUCAACUCAAUUAAAUCUACGAAAUGGAAUUUCAAAUUAAUUC-UUUUCUCCUUUUCCGUUCCAUGAGCUUCAAUUUGCU--- ...(((((((((.(((((((....))))))).)))))))))............(.(((((((..............-..........))))))).)...(((........)))--- ( -20.26, z-score = -1.44, R) >dp4.chrXR_group8 3693224 104 - 9212921 UGAGGGUUGAAAAUAAAUUGCUUUUAAUUUGAUUUCAACUCAAUUAAAUCUACGAAAUGGAAUUUCAAAUUAAUUCUUUUUCUGCUCCUUGGGUGCGU--------CCCUCC---- .(((((((((((.(((((((....))))))).)))))))..............((((.((((((.......)))))).)))).((.((...)).))..--------.)))).---- ( -23.80, z-score = -2.60, R) >droPer1.super_23 1608279 104 - 1662726 UGAGGGUUGAAAAUAAAUUGCUUUUAAUUUGAUUUCAACUCAAUUAAAUCUACGAAAUGGAAUUUCAAAUUAAUUCUUUUUCUGCUCCUUGGGUGCGC--------CCCUCC---- ...(((((((((.(((((((....))))))).)))))))))............((((.((((((.......)))))).))))........(((....)--------))....---- ( -23.90, z-score = -2.47, R) >droWil1.scaffold_180698 327634 109 + 11422946 UGAGGGUUGAAAAUAAAUUGCUUUUAAUUUGAUUUCAACUCAAUUAAAUCUACGAAAUGGAAUUUCAAAUUAAUUC-CUUUCUCUCUAUUUGCUAUAUCU------AUCGGCUGCU ...(((((((((.(((((((....))))))).)))))))))............((((.((((((.......)))))-))))).........(((......------...))).... ( -21.70, z-score = -3.14, R) >droVir3.scaffold_13049 14991488 102 - 25233164 UGAGGGUUGAAAAUAAAUUGCUUUUAAUUUGAUUUCAACUCAAUUAAAUCUACGAAAUGGAAUUUCAAAUUAAUUC-AUUUC--CCGACUUGCCACAUCC-------UUGAU---- ...(((((((((.(((((((....))))))).)))))))))............((((((((............)))-)))))--................-------.....---- ( -18.50, z-score = -1.50, R) >droMoj3.scaffold_6680 2867592 102 + 24764193 UGAGGGUUGAAAAUAAAUUGCUUUUAAUUUGAUUUCAACUCAAUUAAAUCUACGGAAUGGAAUUUGAAAUUAAUUC-AUUUC--UCAACUUGGCACAUCC-------UUGAC---- ((((((((((((.(((((((....))))))).)))))))))............((((((((............)))-)))))--))).(..((.....))-------..)..---- ( -19.20, z-score = -1.65, R) >droGri2.scaffold_15110 19420824 103 - 24565398 UGAGGGUUGAAAAUAAAUUGCUUUUAAUUUGAUUUCAACUCAAUUAAAUCUACGGAAUGGAAUUUCAAAUUAAUUC-AUUAC-ACUGCCUUGGCACAUCC-------UUGAC---- ...(((((((((.(((((((....))))))).)))))))))............(((.(((((((.......)))))-.....-..(((....))))))))-------.....---- ( -22.20, z-score = -2.74, R) >consensus UGAGGGUUGAAAAUAAAUUGCUUUUAAUUUGAUUUCAACUCAAUUAAAUCUACGAAAUGGAAUUUCAAAUUAAUUC_UUUUCUGCUCGCUCGCCUCAC_________UUUGCU___ ...(((((((((.(((((((....))))))).)))))))))............((((..(((((.......)))))..)))).................................. (-16.00 = -16.02 + 0.01)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:58:46 2011