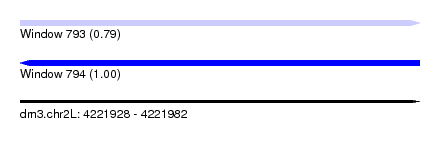
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,221,928 – 4,221,982 |
| Length | 54 |
| Max. P | 0.998050 |

| Location | 4,221,928 – 4,221,982 |
|---|---|
| Length | 54 |
| Sequences | 8 |
| Columns | 54 |
| Reading direction | forward |
| Mean pairwise identity | 94.11 |
| Shannon entropy | 0.11686 |
| G+C content | 0.34102 |
| Mean single sequence MFE | -9.20 |
| Consensus MFE | -6.45 |
| Energy contribution | -6.45 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.791137 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
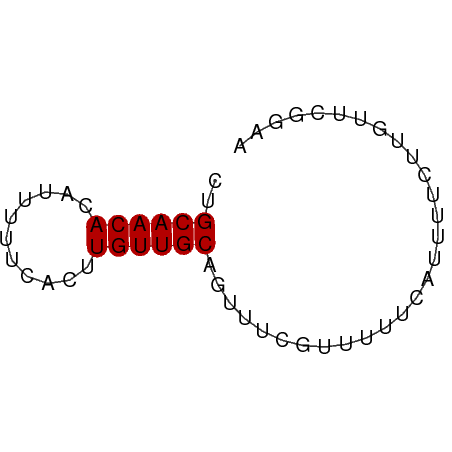
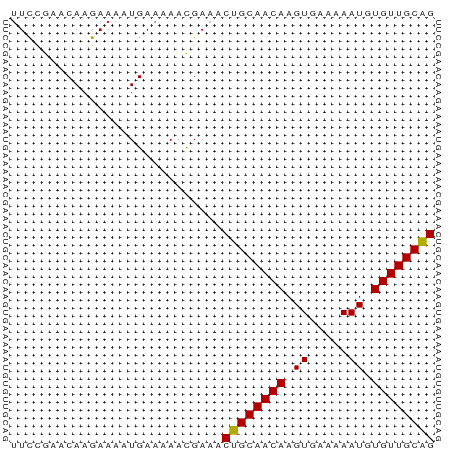
>dm3.chr2L 4221928 54 + 23011544 CUGCAACACAUUUUUCACUUGUUGCAGUUUCGUUUUUCAUUUUCUUGUUCGGAA ((((((((...........))))))))(((((.....((......))..))))) ( -10.40, z-score = -2.69, R) >droSim1.chr2L_random 202837 54 + 909653 CUGCAACACAUUUUUCACUUGUUGCAGUUUCGUUUUUCAUUUUCUUGUUCGGAA ((((((((...........))))))))(((((.....((......))..))))) ( -10.40, z-score = -2.69, R) >droSec1.super_5 2319234 54 + 5866729 CUGCAACACAUUUUUCACUUGUUGCAGUUUCGUUUUUCAUUUUCUUGUUCGGAA ((((((((...........))))))))(((((.....((......))..))))) ( -10.40, z-score = -2.69, R) >dp4.chr4_group2 207929 54 + 1235136 CUGCAACACAUUUUUCACUUGUUGCAGUUUCGUUUUUCAUUUUCUUGUUCGGAA ((((((((...........))))))))(((((.....((......))..))))) ( -10.40, z-score = -2.69, R) >droPer1.super_8 285716 54 - 3966273 CUGCAACACAUUUUUCACUUGUUGCAGUUUCGUUUUUCAUUUUCUUGUUCGGAA ((((((((...........))))))))(((((.....((......))..))))) ( -10.40, z-score = -2.69, R) >droVir3.scaffold_13246 303664 54 + 2672878 CCGCAACAUAUUUUUCACUUGUUGCAGUUUCGUUUUUCAUUUUCUUGUACUAAA ..((((((...........))))))............................. ( -7.20, z-score = -2.60, R) >droMoj3.scaffold_6500 298664 54 + 32352404 CCGCAACAUAUUUUUCACUUGUUGCAGUUUCGUUUUUCAUUUUCUUGUACAGAA ..((((((...........))))))............................. ( -7.20, z-score = -1.81, R) >droGri2.scaffold_15252 215806 53 - 17193109 CCGCAACAUAUUUUUCACUUGUUGCAGUUUCAUUUUUCAUUUUCUUGUACCAA- ..((((((...........))))))............................- ( -7.20, z-score = -3.37, R) >consensus CUGCAACACAUUUUUCACUUGUUGCAGUUUCGUUUUUCAUUUUCUUGUUCGGAA ..((((((...........))))))............................. ( -6.45 = -6.45 + 0.00)

| Location | 4,221,928 – 4,221,982 |
|---|---|
| Length | 54 |
| Sequences | 8 |
| Columns | 54 |
| Reading direction | reverse |
| Mean pairwise identity | 94.11 |
| Shannon entropy | 0.11686 |
| G+C content | 0.34102 |
| Mean single sequence MFE | -11.01 |
| Consensus MFE | -10.87 |
| Energy contribution | -10.64 |
| Covariance contribution | -0.23 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.99 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.24 |
| SVM RNA-class probability | 0.998050 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
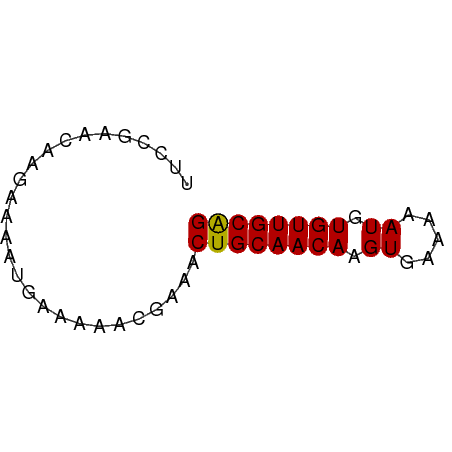
>dm3.chr2L 4221928 54 - 23011544 UUCCGAACAAGAAAAUGAAAAACGAAACUGCAACAAGUGAAAAAUGUGUUGCAG ...........................((((((((..(....)...)))))))) ( -11.20, z-score = -3.20, R) >droSim1.chr2L_random 202837 54 - 909653 UUCCGAACAAGAAAAUGAAAAACGAAACUGCAACAAGUGAAAAAUGUGUUGCAG ...........................((((((((..(....)...)))))))) ( -11.20, z-score = -3.20, R) >droSec1.super_5 2319234 54 - 5866729 UUCCGAACAAGAAAAUGAAAAACGAAACUGCAACAAGUGAAAAAUGUGUUGCAG ...........................((((((((..(....)...)))))))) ( -11.20, z-score = -3.20, R) >dp4.chr4_group2 207929 54 - 1235136 UUCCGAACAAGAAAAUGAAAAACGAAACUGCAACAAGUGAAAAAUGUGUUGCAG ...........................((((((((..(....)...)))))))) ( -11.20, z-score = -3.20, R) >droPer1.super_8 285716 54 + 3966273 UUCCGAACAAGAAAAUGAAAAACGAAACUGCAACAAGUGAAAAAUGUGUUGCAG ...........................((((((((..(....)...)))))))) ( -11.20, z-score = -3.20, R) >droVir3.scaffold_13246 303664 54 - 2672878 UUUAGUACAAGAAAAUGAAAAACGAAACUGCAACAAGUGAAAAAUAUGUUGCGG ...........................((((((((.((.....)).)))))))) ( -10.20, z-score = -2.99, R) >droMoj3.scaffold_6500 298664 54 - 32352404 UUCUGUACAAGAAAAUGAAAAACGAAACUGCAACAAGUGAAAAAUAUGUUGCGG ((((.....))))..............((((((((.((.....)).)))))))) ( -11.70, z-score = -3.48, R) >droGri2.scaffold_15252 215806 53 + 17193109 -UUGGUACAAGAAAAUGAAAAAUGAAACUGCAACAAGUGAAAAAUAUGUUGCGG -..........................((((((((.((.....)).)))))))) ( -10.20, z-score = -3.33, R) >consensus UUCCGAACAAGAAAAUGAAAAACGAAACUGCAACAAGUGAAAAAUGUGUUGCAG ...........................((((((((.((.....)).)))))))) (-10.87 = -10.64 + -0.23)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:15:42 2011