
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 2,837,726 – 2,837,826 |
| Length | 100 |
| Max. P | 0.893987 |

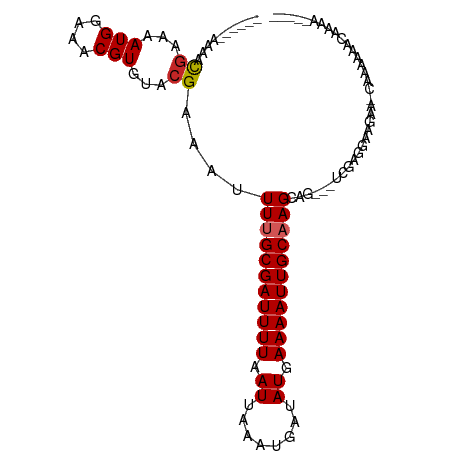
| Location | 2,837,726 – 2,837,820 |
|---|---|
| Length | 94 |
| Sequences | 10 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 82.63 |
| Shannon entropy | 0.34916 |
| G+C content | 0.30981 |
| Mean single sequence MFE | -15.35 |
| Consensus MFE | -11.27 |
| Energy contribution | -11.48 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.893987 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 2837726 94 + 24543557 ------AAAACGAAAAUGGAAACGUGUACGAAAUUUUGCGAUUUUAAUUAAAUGAUAUGAAAAUUGCAAGCAG---UCGAGGAAGAA-CAAAAAAAAAACAAAA- ------.........(((....)))...(((...(((((((((((.((........)).)))))))))))...---)))........-................- ( -15.30, z-score = -2.04, R) >droSim1.chr3L 2391665 90 + 22553184 ------AAAACGAAAAUGGAAACGUGUACGAAAUUUUGCGAUUUUAAUUAAAUGAUAUGAAAAUUGCAAGCAG---UCGAGGAAGAA-CAAAAAACAAAG----- ------.........(((....)))...(((...(((((((((((.((........)).)))))))))))...---)))........-............----- ( -15.30, z-score = -2.21, R) >droSec1.super_2 2858034 96 + 7591821 AAAACGAAAAUGGAAAUGGAAACGUGUACGAAAUUUUGCGAUUUUAAUUAAAUGAUAUGAAAAUUGCAAGCAG---UCGAGGAAGAA-CAAAAAACCAAG----- ..........(((..(((....)))...(((...(((((((((((.((........)).)))))))))))...---)))........-.......)))..----- ( -18.30, z-score = -2.89, R) >droYak2.chr3L 15101968 96 - 24197627 AAAACGAAAACAAAAAUGGAAACGUGUACGAAAUUUUGCGAUUUUAAUUAAAUGAUAUGAAAAUUGCAAGCAG---UCGAGGAAGAA-CAAAAAGCUUGG----- ..........(((..(((....)))...(((...(((((((((((.((........)).)))))))))))...---)))........-........))).----- ( -15.90, z-score = -1.47, R) >droEre2.scaffold_4784 2863673 87 + 25762168 ------AAAACGAAAAUGGAAACGUGUACGAAAUUUUGCGAUUUUAAUUAAAUGAUAUGAAAAUUGCAAGCAG---UCGAGGAAGAA-CAAAAAAGG-------- ------.........(((....)))...(((...(((((((((((.((........)).)))))))))))...---)))........-.........-------- ( -15.30, z-score = -2.31, R) >droAna3.scaffold_13337 14043016 89 - 23293914 ------AAAACGAAAAUGGAAACGUGUACGAAAUUUUGCGAUUUUAAUUAAAUGAUAUGAAAAUUGCAAGCAG---UCGAG-AAGAA-CAAAAAGCGAAG----- ------....((...(((....)))...(((...(((((((((((.((........)).)))))))))))...---)))..-.....-.......))...----- ( -16.00, z-score = -2.02, R) >dp4.chrXR_group8 5541990 99 - 9212921 ------AAAACGAAAAUGGAAACGUGUACGAAAUUUUGCGAUUUUAAUUAAAUGAUAUGAAAAUUGCAAGCAGCAGUCGAGGAAGAAGCGAGAAAAAUAACAGGA ------....((...(((....)))...))....(((((((((((.((........)).)))))))))))..((..((......)).))................ ( -15.40, z-score = -1.07, R) >droPer1.super_23 77761 99 - 1662726 ------AAAACGAAAAUGGAAACGUGUACGAAAUUUUGCGAUUUUAAUUAAAUGAUAUGAAAAUUGCAAGCAGCAGUCGAGGAAGAAGCGAGAAAAAUAACAAGA ------....((...(((....)))...))....(((((((((((.((........)).)))))))))))..((..((......)).))................ ( -15.40, z-score = -1.11, R) >droVir3.scaffold_13049 14968663 95 + 25233164 -------AAACGAAAAUGAAAACGUGUACGAAAUUU-GCGAUUUUAAUUAAAUGAUAUGAAAAUUGCAAGGCGCACCAAAGCGAAAGAGAAAAA-CAACAACAG- -------...............(((((.((...(((-((((((((.((........)).))))))))))).)).))....)))...........-.........- ( -12.80, z-score = -0.93, R) >droGri2.scaffold_15110 19388312 97 + 24565398 ------AAAACGAAAAUGAAAACGUGUACGAAAUUU-GCGAUUUUAAUUAAAUGAUAUGAAAAUUGCAAGGCGCACUAAAACGAAAGAGAAAAAACAAAAAACG- ------................(((((.((...(((-((((((((.((........)).))))))))))).)).)).....(....)..............)))- ( -13.80, z-score = -1.46, R) >consensus ______AAAACGAAAAUGGAAACGUGUACGAAAUUUUGCGAUUUUAAUUAAAUGAUAUGAAAAUUGCAAGCAG___UCGAGGAAGAA_CAAAAAACAAAA_____ ..........((...(((....)))...))....(((((((((((.((........)).)))))))))))................................... (-11.27 = -11.48 + 0.21)


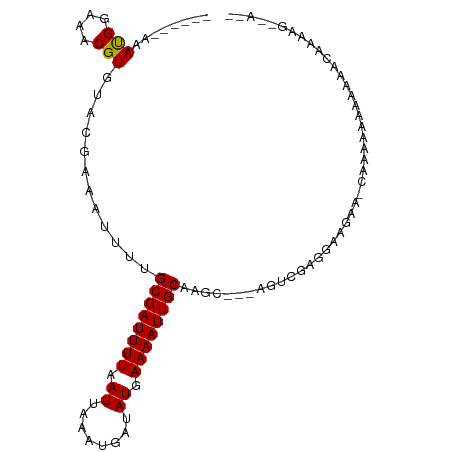
| Location | 2,837,733 – 2,837,826 |
|---|---|
| Length | 93 |
| Sequences | 12 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 77.26 |
| Shannon entropy | 0.46976 |
| G+C content | 0.31429 |
| Mean single sequence MFE | -15.05 |
| Consensus MFE | -8.09 |
| Energy contribution | -8.02 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.94 |
| SVM RNA-class probability | 0.857590 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 2837733 93 + 24543557 ------AAAUGGAAACGUGUACGAAAUUUUGCGAUUUUAAUUAAAUGAUAUGAAAAUUGCAAGC---AGUCGAGGAAGAA--CAAAAAAAAAACAAAACAAAAG ------..(((....)))...(((...(((((((((((.((........)).))))))))))).---..)))........--...................... ( -15.30, z-score = -2.37, R) >droSim1.chr3L 2391672 89 + 22553184 ------AAAUGGAAACGUGUACGAAAUUUUGCGAUUUUAAUUAAAUGAUAUGAAAAUUGCAAGC---AGUCGAGGAAGAA-CAAAAAACAAAGCAAAAG----- ------..(((....)))...(((...(((((((((((.((........)).))))))))))).---..)))........-..................----- ( -15.30, z-score = -2.09, R) >droSec1.super_2 2858041 95 + 7591821 AAAUGGAAAUGGAAACGUGUACGAAAUUUUGCGAUUUUAAUUAAAUGAUAUGAAAAUUGCAAGC---AGUCGAGGAAGAA-CAAAAAACCAAGCAAAAG----- ...(((..(((....)))...(((...(((((((((((.((........)).))))))))))).---..)))........-.......)))........----- ( -18.30, z-score = -2.79, R) >droYak2.chr3L 15101981 89 - 24197627 ------AAAUGGAAACGUGUACGAAAUUUUGCGAUUUUAAUUAAAUGAUAUGAAAAUUGCAAGC---AGUCGAGGAAGAA-CAAAAAGCUUGGCAAAAG----- ------..(((....))).........(((((((((((.((........)).))))))))))).---.((((((......-.......)))))).....----- ( -16.72, z-score = -1.71, R) >droEre2.scaffold_4784 2863680 86 + 25762168 ------AAAUGGAAACGUGUACGAAAUUUUGCGAUUUUAAUUAAAUGAUAUGAAAAUUGCAAGC---AGUCGAGGAAGAA-CAAAAAA---GGCAAAAG----- ------..(((....)))...(((...(((((((((((.((........)).))))))))))).---..)))........-.......---........----- ( -15.30, z-score = -2.20, R) >droAna3.scaffold_13337 14043023 88 - 23293914 ------AAAUGGAAACGUGUACGAAAUUUUGCGAUUUUAAUUAAAUGAUAUGAAAAUUGCAAGC---AGUCGAGAAGAACAAAAAGCGAAGAACAAA------- ------..(((....)))...(((...(((((((((((.((........)).))))))))))).---..))).........................------- ( -15.30, z-score = -1.83, R) >dp4.chrXR_group8 5541997 98 - 9212921 ------AAAUGGAAACGUGUACGAAAUUUUGCGAUUUUAAUUAAAUGAUAUGAAAAUUGCAAGCAGCAGUCGAGGAAGAAGCGAGAAAAAUAACAGGAGAGAAG ------..(((....))).........(((((((((((.((........)).)))))))))))..((..((......)).))...................... ( -13.50, z-score = -0.61, R) >droPer1.super_23 77768 98 - 1662726 ------AAAUGGAAACGUGUACGAAAUUUUGCGAUUUUAAUUAAAUGAUAUGAAAAUUGCAAGCAGCAGUCGAGGAAGAAGCGAGAAAAAUAACAAGAGAGAAG ------..(((....))).........(((((((((((.((........)).)))))))))))..((..((......)).))...................... ( -13.50, z-score = -0.70, R) >droWil1.scaffold_180698 310341 95 - 11422946 ------AAACGAAAACGUGUACCAAAUUU-GCGAUUUUAAUUAAAUGAUAUGAAAAUUGCAAGAAG-AAAAGAAGCAACGAGAAAAGAGAGAAUUUGGUGGAG- ------..(((....))).((((((((((-((((((((.((........)).))))))))......-...........(.......)...))))))))))...- ( -14.80, z-score = -2.41, R) >droVir3.scaffold_13049 14968669 94 + 25233164 ------AAAUGAAAACGUGUACGAAAUUU-GCGAUUUUAAUUAAAUGAUAUGAAAAUUGCAAGGCGCACCAAA-GCGAAAGAGAAAAACAACAACA-GCAAAA- ------...((......(((......(((-((((((((.((........)).))))))))))).(((......-)))..........)))......-.))...- ( -13.02, z-score = -0.49, R) >droMoj3.scaffold_6680 2842710 95 - 24764193 ------AAAUGAAAACGUGUACGAAAUUU-GCGAUUUUAAUUAAAUGAUAUGAAAAUUGCAAGGAGCACCAAAAGCGAACAACAAGAAGAAGACGAUGCAAA-- ------...........((((((...(((-((((((((.((........)).)))))))))))..((.......)).................)).))))..-- ( -13.50, z-score = -1.20, R) >droGri2.scaffold_15110 19388319 96 + 24565398 ------AAAUGAAAACGUGUACGAAAUUU-GCGAUUUUAAUUAAAUGAUAUGAAAAUUGCAAGGCGCACUAAA-ACGAAAGAGAAAAAACAAAAAACGUGAAAA ------........((((((.((...(((-((((((((.((........)).))))))))))).)).))....-.(....)..............))))..... ( -16.00, z-score = -2.17, R) >consensus ______AAAUGGAAACGUGUACGAAAUUUUGCGAUUUUAAUUAAAUGAUAUGAAAAUUGCAAGC___AGUCGAGGAAGAA_CAAAAAAAAAAACAAAAG__A__ ........(((....)))............((((((((.((........)).))))))))............................................ ( -8.09 = -8.02 + -0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:58:42 2011