
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 2,817,667 – 2,817,759 |
| Length | 92 |
| Max. P | 0.993113 |

| Location | 2,817,667 – 2,817,759 |
|---|---|
| Length | 92 |
| Sequences | 10 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 76.74 |
| Shannon entropy | 0.48478 |
| G+C content | 0.37025 |
| Mean single sequence MFE | -20.28 |
| Consensus MFE | -13.87 |
| Energy contribution | -13.66 |
| Covariance contribution | -0.21 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.993113 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr3L 2817667 92 + 24543557 AAAUUAAAUUGCGCAGAUAAAUUUGCGCAACGCUAAUAAAUUAAUAUUUAUGGUUGCCUGCCC---UGCAUAAUUGUAAUUCUGGCUUCC-----CGCCU- ........(((((((((....))))))))).((..((((((....))))))((..(((.....---((((....)))).....)))..))-----.))..- ( -22.40, z-score = -2.21, R) >droSim1.chr3L 2370171 92 + 22553184 AAAUUAAAUUGCGCAGAUAAAUUUGCGCAACGCUAAUAAAUUAAUAUUUAUGGUUGCCUGCCC---UGCAUAAUUGUAAUUCUGGCUUCC-----CGCCU- ........(((((((((....))))))))).((..((((((....))))))((..(((.....---((((....)))).....)))..))-----.))..- ( -22.40, z-score = -2.21, R) >droSec1.super_2 2838122 92 + 7591821 AAAUUAAAUUGCGCAGAUAAAUUUGCGCAACGCUAAUAAAUUAAUAUUUAUGGUGGCCUGCCC---UGCAUAAUUGUAAUUCUGGCUUCC-----CGCCU- ........(((((((((....))))))))).....((((((....))))))(((((...(((.---((((....)))).....)))...)-----)))).- ( -25.70, z-score = -2.76, R) >droYak2.chr3L 15080974 92 - 24197627 AAAUUAAAUUGCGCAGAUAAAUUUGCGCAACGCUAAUAAAUUAAUAUUUAUGGUUGCCUGCCC---UGCAUAAUUGUAAUUCUCGCUUUC-----CGCCU- ........(((((((((....))))))))).((..((((((....))))))((..((..(...---((((....))))...)..))...)-----)))..- ( -18.90, z-score = -1.18, R) >droEre2.scaffold_4784 2843215 92 + 25762168 AAAUUAAAUUGCGCAGAUAAAUUUGCGCAACGCUAAUAAAUUAAUAUUUAUGGUUGCCUGCCC---UGCAUAAUUGUAAUUCUCGCUUCC-----CGCCU- ........(((((((((....))))))))).((..((((((....))))))((..((..(...---((((....))))...)..))..))-----.))..- ( -18.30, z-score = -1.39, R) >droAna3.scaffold_13337 14023323 97 - 23293914 AAAUUAAAUUGCGCAGAUAAAUUUGCGCAACGCUAAUAAAUUAAUAUUUAUGGUUGCCUGCCC---AGCAUAAUUGUAAUUCUGACUUCCUAGGCCGCCU- ........(((((((((....))))))))).....((((((....))))))(((.(((((...---((((.(((....))).)).))...))))).))).- ( -23.30, z-score = -1.94, R) >dp4.chrXR_group8 3645627 86 + 9212921 AAAUUAAAUUGCGCAGAUAAAUUUGCGCAACGCUAAUAAAUUAAUAUUUAUGGUUGCAUGCCC---UGCAUAAUUGUCUCCCCCUCCCC------------ .(((((..(((((((((....))))))))).((..((((((....))))))(((.....))).---.)).)))))..............------------ ( -16.70, z-score = -2.10, R) >droVir3.scaffold_13049 14944041 82 + 25233164 AAAUUUAAUUGCGCAGAUAAAUUUGCGCAAUGCUAAUAAAUUAAUAUUUAUGUGCGUGCAUGCCCUUCCCCAUUCCCUAGCC------------------- .......((((((((((....))))))))))((((((((((....))))))(((....)))................)))).------------------- ( -17.60, z-score = -2.60, R) >droMoj3.scaffold_6680 2815134 76 - 24764193 AAAUUUAAUUGCGCAGAUAAAUUUGCGCAAUGCUAAUAAAUUAAUAUUUAUGUGCGUGCAUU----CCCUUAACUUACCC--------------------- .......((((((((((....))))))))))((..((((((....))))))..)).......----..............--------------------- ( -16.20, z-score = -2.39, R) >droGri2.scaffold_15110 19361760 101 + 24565398 AAAUUUAAUUGCGCAGAUAAAUUUGCGCAAUGCUAAUAAAUUAAUAUUUAUGUGCGUGCGCUUGUACCCUUAAUAUGGAUACACCCCUCCCCCCACCCGCA .........((((...........(((((.(((..((((((....))))))..)))))))).((((.((.......)).))))..............)))) ( -21.30, z-score = -2.40, R) >consensus AAAUUAAAUUGCGCAGAUAAAUUUGCGCAACGCUAAUAAAUUAAUAUUUAUGGUUGCCUGCCC___UGCAUAAUUGUAAUUCUCGCUUCC_____CGCCU_ ........(((((((((....))))))))).((..((((((....))))))....))............................................ (-13.87 = -13.66 + -0.21)

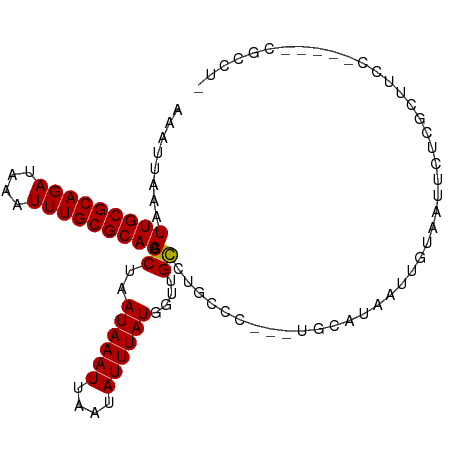
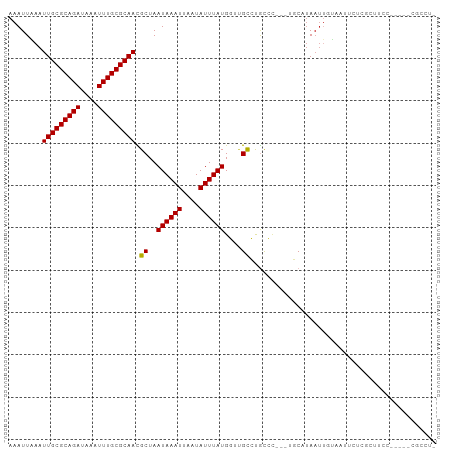
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:58:39 2011