
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 2,816,962 – 2,817,063 |
| Length | 101 |
| Max. P | 0.834930 |

| Location | 2,816,962 – 2,817,061 |
|---|---|
| Length | 99 |
| Sequences | 9 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 69.68 |
| Shannon entropy | 0.59480 |
| G+C content | 0.45468 |
| Mean single sequence MFE | -28.11 |
| Consensus MFE | -12.15 |
| Energy contribution | -12.06 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.50 |
| SVM RNA-class probability | 0.721808 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
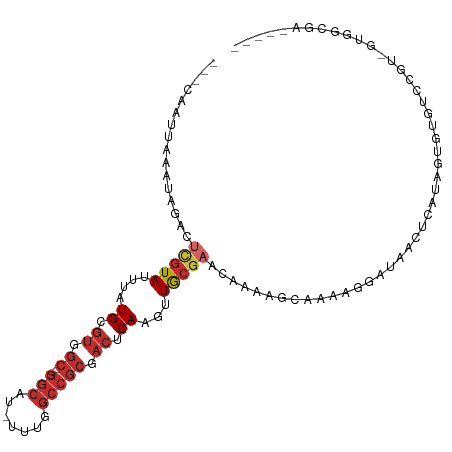
>dm3.chr3L 2816962 99 - 24543557 ---CAAUUAAAUAGACUCGUAUUUAUGCGUGGC---GGCAU-UUUGGCCGCGACUCAAGUUGCGAACAAAAGCAAAAGGAUAACUCAUAGUGUGUCCGU-GCGGCGA----- ---....((((((......))))))..(((.((---(((.(-((((..((((((....))))))..)))))))....(((((((.....)).))))).)-)).))).----- ( -29.70, z-score = -1.24, R) >droSim1.chr3L 2369495 99 - 22553184 ---CAAUUAAAUAGACUCGUAUUUAUGCGUGGC---GGCAU-UUUGGCCGCGACUCAAGUUGCGAACAAAAGCAAAAGGAUAACUCAUAGUGUGUCCGU-GUGGCGA----- ---....((((((......))))))..(((.((---(((.(-((((..((((((....))))))..)))))))....(((((((.....)).))))).)-)).))).----- ( -28.20, z-score = -0.98, R) >droSec1.super_2 2837480 99 - 7591821 ---CAUUUAAAUAGACUCGUAUUUAUGCGUGGC---GGCAU-UUUGGCCGCGACUCAAGUUGCGAACAAAAGCAAAAGGAUAACUCAUAGUGUGUCCGU-GUGGCGA----- ---....((((((......))))))..(((.((---(((.(-((((..((((((....))))))..)))))))....(((((((.....)).))))).)-)).))).----- ( -28.20, z-score = -0.93, R) >droYak2.chr3L 15080271 99 + 24197627 ---CAAUUAAAUAGACUCGUAUUUAUGCGUGGC---GGCAU-UUUGGCCGCGACUCAAGUUGCGAACAAAAGCAGAAGGAUAACUCAUAGUGUGUCCGU-GUGGCAA----- ---............(.((((....)))).)((---.((((-((((..((((((....))))))..)))).......(((((((.....)).)))))))-)).))..----- ( -27.50, z-score = -0.83, R) >droEre2.scaffold_4784 2842502 99 - 25762168 ---CAAUUAAAUAGACUCGUAUUUAUGCGUGGC---GGCAU-UUUGGCCGCGACUCAAGUUGCGAACAAAAGCAGAAGGAUAACUCAUAGUGUGUCCGU-GUGGCAA----- ---............(.((((....)))).)((---.((((-((((..((((((....))))))..)))).......(((((((.....)).)))))))-)).))..----- ( -27.50, z-score = -0.83, R) >droAna3.scaffold_13337 14022904 108 + 23293914 ---CCCUUAAAUAGUGUCGUAUUUAUGCGUGGC-AUGGCGUAUUUGGCCGCGACUCAAGUUGCACACACAAACACAAAGGCGAAAAAGGAUAACUCUGUCCUGGCGCGUCGG ---.((.......((((.(((....)))(((..-..(((.......)))(((((....))))).)))....))))...((((....((((((....))))))....)))))) ( -31.00, z-score = -0.54, R) >droPer1.super_23 1559851 95 - 1662726 CUCCCAAUAUACACGUUCGUAUUUAUGCGUAGC---GGCGUAUUUGGCCGCGACUCAAGUUGCGAACAAAAGCCAAAAGGAUAUCCAGGAGGGGGAAA-------------- (((((.........(((((((....)))).)))---(((...((((..((((((....))))))..)))).)))....((....))....)))))...-------------- ( -32.10, z-score = -2.78, R) >dp4.chrXR_group8 3645265 95 - 9212921 CUCCCAAUAUACACGUUCGUAUUUAUGCGUAGC---GGCGUAUUUGGCCGCGACUCAAGUUGCGAACAAAAGCCAAAAGGAUAUCCAGGAGGGGGAAA-------------- (((((.........(((((((....)))).)))---(((...((((..((((((....))))))..)))).)))....((....))....)))))...-------------- ( -32.10, z-score = -2.78, R) >droWil1.scaffold_180698 287602 88 + 11422946 ----------AUGUACAUAUAUUUAUGUAAAAAAACCAACUCUUUGGCCGCGACUCAAGUUACGCACAGAAUAAGGAUAAUGUGUGAAUGAAAGAGAA-------------- ----------...((((((....))))))..........((((((.(....(((....))).((((((..((....))..))))))..).))))))..-------------- ( -16.70, z-score = -1.49, R) >consensus ___CAAUUAAAUAGACUCGUAUUUAUGCGUGGC___GGCAU_UUUGGCCGCGACUCAAGUUGCGAACAAAAGCAAAAGGAUAACUCAUAGUGUGUCCGU_GUGGCGA_____ .................((((....))))........((...((((..((((((....))))))..)))).))....................................... (-12.15 = -12.06 + -0.10)


| Location | 2,816,964 – 2,817,063 |
|---|---|
| Length | 99 |
| Sequences | 9 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 69.81 |
| Shannon entropy | 0.59282 |
| G+C content | 0.46524 |
| Mean single sequence MFE | -28.33 |
| Consensus MFE | -13.29 |
| Energy contribution | -13.47 |
| Covariance contribution | 0.18 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.834930 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
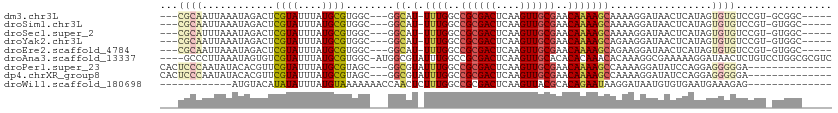
>dm3.chr3L 2816964 99 - 24543557 ---CGCAAUUAAAUAGACUCGUAUUUAUGCGUGGC---GGCAU-UUUGGCCGCGACUCAAGUUGCGAACAAAAGCAAAAGGAUAACUCAUAGUGUGUCCGU-GCGGC----- ---((((((((((((......))))).((.((.((---(((..-....))))).)).))))))))).......(((...(((((((.....)).))))).)-))...----- ( -31.30, z-score = -1.69, R) >droSim1.chr3L 2369497 99 - 22553184 ---CGCAAUUAAAUAGACUCGUAUUUAUGCGUGGC---GGCAU-UUUGGCCGCGACUCAAGUUGCGAACAAAAGCAAAAGGAUAACUCAUAGUGUGUCCGU-GUGGC----- ---((((((((((((......))))).((.((.((---(((..-....))))).)).))))))))).......(((...(((((((.....)).))))).)-))...----- ( -29.00, z-score = -1.19, R) >droSec1.super_2 2837482 99 - 7591821 ---CGCAUUUAAAUAGACUCGUAUUUAUGCGUGGC---GGCAU-UUUGGCCGCGACUCAAGUUGCGAACAAAAGCAAAAGGAUAACUCAUAGUGUGUCCGU-GUGGC----- ---(((((..........(((((....((.((.((---(((..-....))))).)).))...)))))............(((((((.....)).)))))))-)))..----- ( -28.50, z-score = -0.98, R) >droYak2.chr3L 15080273 99 + 24197627 ---CGCAAUUAAAUAGACUCGUAUUUAUGCGUGGC---GGCAU-UUUGGCCGCGACUCAAGUUGCGAACAAAAGCAGAAGGAUAACUCAUAGUGUGUCCGU-GUGGC----- ---.(((..((((((......)))))))))((.((---(((.(-((((..((((((....))))))..)))))))....(((((((.....)).))))).)-)).))----- ( -28.20, z-score = -0.72, R) >droEre2.scaffold_4784 2842504 99 - 25762168 ---CGCAAUUAAAUAGACUCGUAUUUAUGCGUGGC---GGCAU-UUUGGCCGCGACUCAAGUUGCGAACAAAAGCAGAAGGAUAACUCAUAGUGUGUCCGU-GUGGC----- ---.(((..((((((......)))))))))((.((---(((.(-((((..((((((....))))))..)))))))....(((((((.....)).))))).)-)).))----- ( -28.20, z-score = -0.72, R) >droAna3.scaffold_13337 14022906 107 + 23293914 ----GCCCUUAAAUAGUGUCGUAUUUAUGCGUGGC-AUGGCGUAUUUGGCCGCGACUCAAGUUGCACACACAAACACAAAGGCGAAAAAGGAUAACUCUGUCCUGGCGCGUC ----...........((((.(((....)))(((..-..(((.......)))(((((....))))).)))....))))....(((....((((((....))))))..)))... ( -29.00, z-score = -0.04, R) >droPer1.super_23 1559853 95 - 1662726 CACUCCCAAUAUACACGUUCGUAUUUAUGCGUAGC---GGCGUAUUUGGCCGCGACUCAAGUUGCGAACAAAAGCCAAAAGGAUAUCCAGGAGGGGGA-------------- ..(((((.........(((((((....)))).)))---(((...((((..((((((....))))))..)))).)))....((....))....))))).-------------- ( -32.60, z-score = -2.87, R) >dp4.chrXR_group8 3645267 95 - 9212921 CACUCCCAAUAUACACGUUCGUAUUUAUGCGUAGC---GGCGUAUUUGGCCGCGACUCAAGUUGCGAACAAAAGCCAAAAGGAUAUCCAGGAGGGGGA-------------- ..(((((.........(((((((....)))).)))---(((...((((..((((((....))))))..)))).)))....((....))....))))).-------------- ( -32.60, z-score = -2.87, R) >droWil1.scaffold_180698 287604 86 + 11422946 ------------AUGUACAUAUAUUUAUGUAAAAAAACCAACUCUUUGGCCGCGACUCAAGUUACGCACAGAAUAAGGAUAAUGUGUGAAUGAAAGAG-------------- ------------...((((((....))))))..........((((((.(....(((....))).((((((..((....))..))))))..).))))))-------------- ( -15.60, z-score = -1.19, R) >consensus ___CGCAAUUAAAUAGACUCGUAUUUAUGCGUGGC___GGCAU_UUUGGCCGCGACUCAAGUUGCGAACAAAAGCAAAAGGAUAACUCAUAGUGUGUCCGU_GUGGC_____ ...((((............((((....))))........((...((((..((((((....))))))..)))).)).................))))................ (-13.29 = -13.47 + 0.18)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:58:38 2011