
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 2,805,863 – 2,805,993 |
| Length | 130 |
| Max. P | 0.993092 |

| Location | 2,805,863 – 2,805,958 |
|---|---|
| Length | 95 |
| Sequences | 9 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 82.41 |
| Shannon entropy | 0.31402 |
| G+C content | 0.42518 |
| Mean single sequence MFE | -31.29 |
| Consensus MFE | -21.23 |
| Energy contribution | -21.39 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.993092 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 2805863 95 + 24543557 ----------------------UGCUUGUAUUUGGCGCGCUGUGUGUGCUUUGCGUUUAUUGAACUCGCGACGCGGCAUGAGGUUAAUUGACCUCGUGCUUAAUGCAAAAAUAAAUA ----------------------...(((((((.((((((((((((.(((....((.....)).....))))))))))..((((((....))))))))))).)))))))......... ( -34.30, z-score = -3.22, R) >droSim1.chr3L 2358453 95 + 22553184 ----------------------UGCUUGUAUUUGGCGCGCUGUGUGUGCUUUGCGUUUAUUGAACUCGCGACGCGGCAUGAGGUUAAUUGACCUCGUGCUUAAUGCAAAAAUAAAUA ----------------------...(((((((.((((((((((((.(((....((.....)).....))))))))))..((((((....))))))))))).)))))))......... ( -34.30, z-score = -3.22, R) >droSec1.super_2 2826459 95 + 7591821 ----------------------UGCUUGUAUUUGGCGCGCUGUGUGUGCUUUGCGAUUAUUGAACUCGCGACGCGGCAUGAGGUUAAUUGACCUCGUGCUUAAUGCAAAAAUAAAUA ----------------------...(((((((.((((((((((((......(((((.........))))))))))))..((((((....))))))))))).)))))))......... ( -35.00, z-score = -3.60, R) >droYak2.chr3L 15068916 95 - 24197627 ----------------------UGCUUGUGUUUGGCGCGCUGUGUGUGCUUUGCGUUUAUUGAACUCGCGACGCGGCAUGAGGUUAAUUGACCUCGUGCUUAAUGCAAAAAUAAAUA ----------------------...(((((((.((((((((((((.(((....((.....)).....))))))))))..((((((....))))))))))).)))))))......... ( -33.30, z-score = -2.68, R) >droEre2.scaffold_4784 2831373 95 + 25762168 ----------------------UGCUUGUGUUUGGCGCGCUGUAUGUGCUUUGCGUUUAUUGAACUUGCGACGCGGCAUGAGGUUAAUUGACCUCGUGCUUAAUGCAAAAAUAAAUA ----------------------.......((..((((((.....))))))..))(((((((....(((((....(((((((((((....)))))))))))...))))).))))))). ( -32.10, z-score = -2.72, R) >droAna3.scaffold_13337 14012189 117 - 23293914 UGUUUAGUUUUUUCGCCUUCUGCUCCACUGUGUGUGUGUGAGUGUGUGCUUUGCGUUUAUUGAACUCCCGUCGCGGCAUGAGGUUAAUUGACCUCGUGCUUAAUGCAAAAAUAAAUA .(((((((.....(((...(((((((((.......))).))))).)......)))...))))))).......(((((((((((((....)))))))))))....))........... ( -31.40, z-score = -2.53, R) >droMoj3.scaffold_6680 2796634 92 - 24764193 -------------------------UAGUGUGUGUGUGUGUGUGAUUGCUUAGCGUUUGUUGAACUCGCCUUGUGGCAUGAGGUUAAUUGACCUCGUGCUUAAUGCAAAAAUAAAUA -------------------------...((((((((.((..(..(.(((...))).)..)...)).))).....(((((((((((....)))))))))))..))))).......... ( -23.70, z-score = -1.95, R) >droVir3.scaffold_13049 14927888 92 + 25233164 -------------------------UAGUCUGUGUGUGUGUGUGUGUGCUUAGCGUUUGUUGAACUCGCGCUGUGGCAUGAGGUUAAUUGACCUCGUGCUUAAUGCAAAAAUAAAUA -------------------------.....(((...((((((((((.(.(((((....))))).).)))))...(((((((((((....)))))))))))..)))))...))).... ( -28.70, z-score = -3.16, R) >droGri2.scaffold_15110 19345616 91 + 24565398 --------------------------UAUAUAUAUUUGUUAGUGUGUGCUUUGCGUUUGUUGAACUCGCGCUGUGGCAUGAGGUUAAUUGACCUCGUGCUUAAUGCAAAAAUAAAUA --------------------------......((((((((..(((((((...((((..(.....)..)))).))(((((((((((....)))))))))))..)))))..)))))))) ( -28.80, z-score = -3.81, R) >consensus ______________________UGCUUGUAUUUGGCGCGCUGUGUGUGCUUUGCGUUUAUUGAACUCGCGACGCGGCAUGAGGUUAAUUGACCUCGUGCUUAAUGCAAAAAUAAAUA .................................................((((((((.........(((...)))((((((((((....))))))))))..))))))))........ (-21.23 = -21.39 + 0.16)
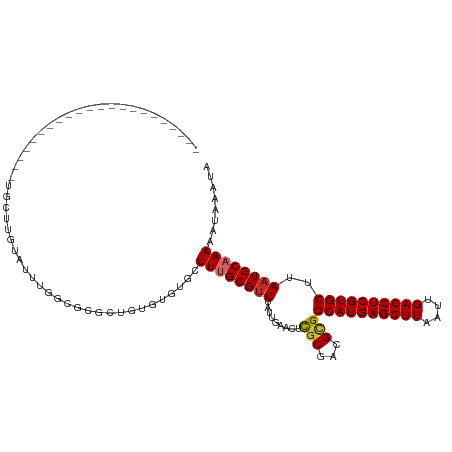
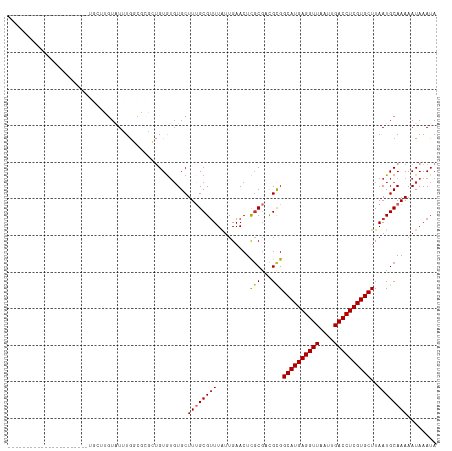
| Location | 2,805,863 – 2,805,958 |
|---|---|
| Length | 95 |
| Sequences | 9 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 82.41 |
| Shannon entropy | 0.31402 |
| G+C content | 0.42518 |
| Mean single sequence MFE | -20.57 |
| Consensus MFE | -14.24 |
| Energy contribution | -14.47 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.939889 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 2805863 95 - 24543557 UAUUUAUUUUUGCAUUAAGCACGAGGUCAAUUAACCUCAUGCCGCGUCGCGAGUUCAAUAAACGCAAAGCACACACAGCGCGCCAAAUACAAGCA---------------------- ..........(((.....(((.(((((......))))).))).(((.(((..(((.....)))((...)).......)))))).........)))---------------------- ( -21.60, z-score = -1.52, R) >droSim1.chr3L 2358453 95 - 22553184 UAUUUAUUUUUGCAUUAAGCACGAGGUCAAUUAACCUCAUGCCGCGUCGCGAGUUCAAUAAACGCAAAGCACACACAGCGCGCCAAAUACAAGCA---------------------- ..........(((.....(((.(((((......))))).))).(((.(((..(((.....)))((...)).......)))))).........)))---------------------- ( -21.60, z-score = -1.52, R) >droSec1.super_2 2826459 95 - 7591821 UAUUUAUUUUUGCAUUAAGCACGAGGUCAAUUAACCUCAUGCCGCGUCGCGAGUUCAAUAAUCGCAAAGCACACACAGCGCGCCAAAUACAAGCA---------------------- ..........(((.....(((.(((((......))))).))).((((.((((.........))))...((.......)))))).........)))---------------------- ( -23.20, z-score = -2.20, R) >droYak2.chr3L 15068916 95 + 24197627 UAUUUAUUUUUGCAUUAAGCACGAGGUCAAUUAACCUCAUGCCGCGUCGCGAGUUCAAUAAACGCAAAGCACACACAGCGCGCCAAACACAAGCA---------------------- ..........(((.....(((.(((((......))))).))).(((.(((..(((.....)))((...)).......)))))).........)))---------------------- ( -21.60, z-score = -1.57, R) >droEre2.scaffold_4784 2831373 95 - 25762168 UAUUUAUUUUUGCAUUAAGCACGAGGUCAAUUAACCUCAUGCCGCGUCGCAAGUUCAAUAAACGCAAAGCACAUACAGCGCGCCAAACACAAGCA---------------------- ..........(((.....(((.(((((......))))).))).(((.(((..(((.....)))((...)).......)))))).........)))---------------------- ( -21.60, z-score = -2.03, R) >droAna3.scaffold_13337 14012189 117 + 23293914 UAUUUAUUUUUGCAUUAAGCACGAGGUCAAUUAACCUCAUGCCGCGACGGGAGUUCAAUAAACGCAAAGCACACACUCACACACACACAGUGGAGCAGAAGGCGAAAAAACUAAACA ........(((((.....(((.(((((......))))).)))...(((....)))........)))))((.....(((.(((.......))))))......)).............. ( -24.10, z-score = -1.98, R) >droMoj3.scaffold_6680 2796634 92 + 24764193 UAUUUAUUUUUGCAUUAAGCACGAGGUCAAUUAACCUCAUGCCACAAGGCGAGUUCAACAAACGCUAAGCAAUCACACACACACACACACUA------------------------- .........((((.(((.((..(((((......))))).((((....))))............)))))))))....................------------------------- ( -17.70, z-score = -3.69, R) >droVir3.scaffold_13049 14927888 92 - 25233164 UAUUUAUUUUUGCAUUAAGCACGAGGUCAAUUAACCUCAUGCCACAGCGCGAGUUCAACAAACGCUAAGCACACACACACACACACAGACUA------------------------- ..........(((.....(((.(((((......))))).)))...((((.............))))..))).....................------------------------- ( -15.92, z-score = -2.43, R) >droGri2.scaffold_15110 19345616 91 - 24565398 UAUUUAUUUUUGCAUUAAGCACGAGGUCAAUUAACCUCAUGCCACAGCGCGAGUUCAACAAACGCAAAGCACACACUAACAAAUAUAUAUA-------------------------- .......((((((.....(((.(((((......))))).)))...(((....)))........))))))......................-------------------------- ( -17.80, z-score = -3.23, R) >consensus UAUUUAUUUUUGCAUUAAGCACGAGGUCAAUUAACCUCAUGCCGCGUCGCGAGUUCAAUAAACGCAAAGCACACACAGCGCGCCAAACACAAGCA______________________ ..........(((.....(((.(((((......))))).)))......(((...........)))...))).............................................. (-14.24 = -14.47 + 0.22)

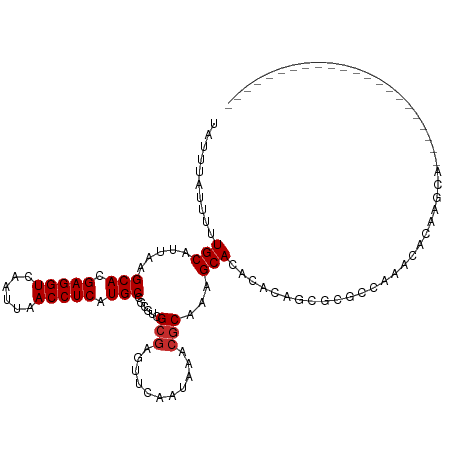
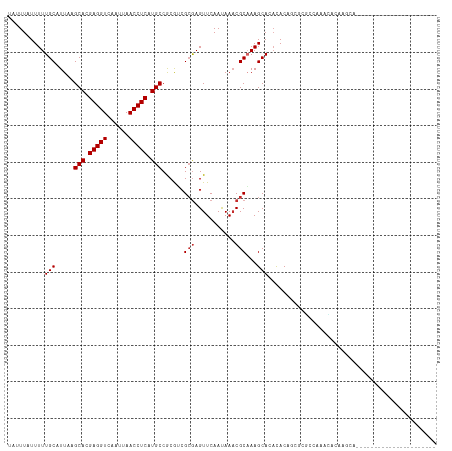
| Location | 2,805,882 – 2,805,993 |
|---|---|
| Length | 111 |
| Sequences | 7 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.86 |
| Shannon entropy | 0.28374 |
| G+C content | 0.39851 |
| Mean single sequence MFE | -29.23 |
| Consensus MFE | -22.12 |
| Energy contribution | -22.61 |
| Covariance contribution | 0.49 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.76 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.957619 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
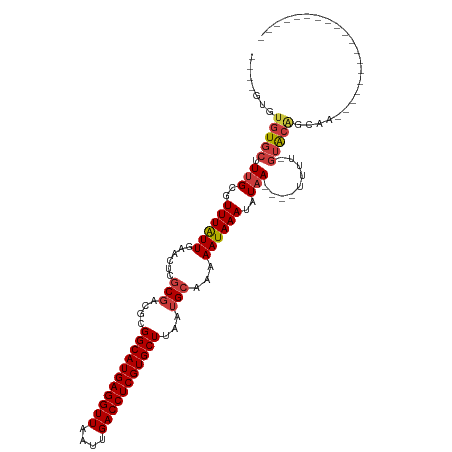
>dm3.chr3L 2805882 111 + 24543557 ----GUGUGUGCUUUGCGUUUAUUGAACUCGCGACGCGGCAUGAGGUUAAUUGACCUCGUGCUUAAUGCAAAAAUAAAUAUAA----UUUU-GUACAGCAACGGCAGCAGCAACAACAAC ----.(((.((((((((((((...))))...((..(((((((((((((....)))))))))))...(((((((.((....)).----))))-)))..))..)))))).))))...))).. ( -34.30, z-score = -2.03, R) >droSim1.chr3L 2358472 108 + 22553184 ----GUGUGUGCUUUGCGUUUAUUGAACUCGCGACGCGGCAUGAGGUUAAUUGACCUCGUGCUUAAUGCAAAAAUAAAUAUAA----UUUU-GUACAGCAACGGCAGCAGCAACAAC--- ----.(((.((((((((((((...))))...((..(((((((((((((....)))))))))))...(((((((.((....)).----))))-)))..))..)))))).)))))))..--- ( -33.30, z-score = -1.84, R) >droYak2.chr3L 15068935 92 - 24197627 ----GUGUGUGCUUUGCGUUUAUUGAACUCGCGACGCGGCAUGAGGUUAAUUGACCUCGUGCUUAAUGCAAAAAUAAAUAUAA----UUUU-GUGCGGCAA------------------- ----.....(((.(((((...........)))))((((((((((((((....)))))))))))....((((((.((....)).----))))-)))))))).------------------- ( -29.40, z-score = -2.38, R) >droEre2.scaffold_4784 2831392 92 + 25762168 ----GUAUGUGCUUUGCGUUUAUUGAACUUGCGACGCGGCAUGAGGUUAAUUGACCUCGUGCUUAAUGCAAAAAUAAAUAUAA----UUUU-GUGCGGCAA------------------- ----((.((..(.(((..((((((....(((((....(((((((((((....)))))))))))...))))).))))))..)))----....-)..))))..------------------- ( -28.20, z-score = -2.46, R) >droAna3.scaffold_13337 14012226 97 - 23293914 GUGAGUGUGUGCUUUGCGUUUAUUGAACUCCCGUCGCGGCAUGAGGUUAAUUGACCUCGUGCUUAAUGCAAAAAUAAAUAUAA----UUUUUGAACAGAAA------------------- .....(((((..((((((((....((.......))..(((((((((((....))))))))))).)))))))).....))))).----..............------------------- ( -22.50, z-score = -1.26, R) >droMoj3.scaffold_6680 2796650 97 - 24764193 ----GUGAUUGCUUAGCGUUUGUUGAACUCGCCUUGUGGCAUGAGGUUAAUUGACCUCGUGCUUAAUGCAAAAAUAAAUAUAAGUUUUUUUUUUACAGCAG------------------- ----((((..(((((..(((((((......((.(((.(((((((((((....)))))))))))))).))...))))))).))))).......)))).....------------------- ( -26.50, z-score = -2.52, R) >droGri2.scaffold_15110 19345631 94 + 24565398 ----GUGUGUGCUUUGCGUUUGUUGAACUCGCGCUGUGGCAUGAGGUUAAUUGACCUCGUGCUUAAUGCAAAAAUAAAUAUAA---UUUUUUGUACAGCAG------------------- ----(((.((.....((....))...)).)))((((((((((((((((....)))))))))))....(((((((.........---.))))))))))))..------------------- ( -30.40, z-score = -3.23, R) >consensus ____GUGUGUGCUUUGCGUUUAUUGAACUCGCGACGCGGCAUGAGGUUAAUUGACCUCGUGCUUAAUGCAAAAAUAAAUAUAA____UUUU_GUACAGCAA___________________ .........((((.(((((((...))))..(((....(((((((((((....)))))))))))...))).......................))).)))).................... (-22.12 = -22.61 + 0.49)

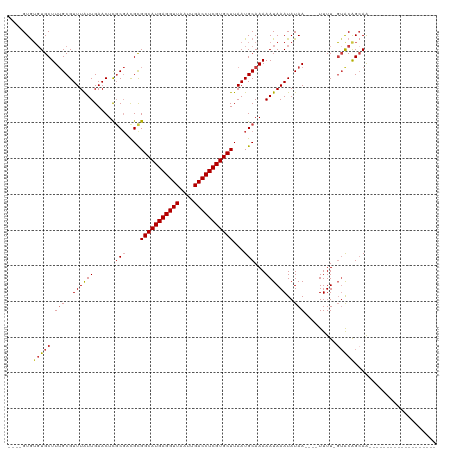
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:58:34 2011