
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,211,219 – 4,211,335 |
| Length | 116 |
| Max. P | 0.744918 |

| Location | 4,211,219 – 4,211,309 |
|---|---|
| Length | 90 |
| Sequences | 10 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 61.13 |
| Shannon entropy | 0.78307 |
| G+C content | 0.47493 |
| Mean single sequence MFE | -23.97 |
| Consensus MFE | -7.52 |
| Energy contribution | -7.63 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.31 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.571714 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 4211219 90 - 23011544 CCAACAAUGCACGCAUGCAAAUGUGUGUAGUUCCAUUUCCAGUUCCAGUUUCGUGCAA---AAGGCUGCAGCGGAAAAAGGUA-GACAAAACAA---- .......(((((((((....)))))))))(((((.(((((.(((.((((((.......---.)))))).))))))))..))..-))).......---- ( -24.00, z-score = -1.53, R) >droPer1.super_8 274667 91 + 3966273 CCAACAAUGUACGCAUGCAAAUGUGCGCG-------CUGUAGUUCCAGUUUCGUGCAGCCAGCAGCAGCAACUUCAUGGGGCACGAAGCAACGUUUAA ...(((.((..(((((....)))))..))-------.))).......(((((((((..(((..((......))...))).)))))))))......... ( -27.50, z-score = -0.08, R) >dp4.chr4_group2 197030 91 - 1235136 CCAACAAUGUACGCAUGCAAAUGUGCGCG-------CUGUAGUUCUAGUUUCGUGCAGCCAGCAGCAGCAACUUCAUGGGGCACGAAGCAACGUUUAA ...(((.((..(((((....)))))..))-------.))).......(((((((((..(((..((......))...))).)))))))))......... ( -27.50, z-score = -0.06, R) >droAna3.scaffold_12916 14632965 81 + 16180835 CCAACAAUUCACGCAUGCAAAUGUGUGUGG------AUCCAGUUCCAGUUUCGUGCAC---GAAUCCGC-GUUUCCAAGGAUG-GCACCUCU------ ..(((....(((((((....)))))))(((------(......)))))))..((((..---..((((..-........)))).-))))....------ ( -21.70, z-score = -0.99, R) >droEre2.scaffold_4929 4284569 82 - 26641161 CCAACAAUGCACGCAUGCAAAUGUGUGUAG------UUCCAGUUCCAGUUUCGUGCAG---AAAGCUGAAGCGGAAAAAAGCU-GAAAAAAA------ .......(((((((((....))))))))).------...(((((((.((((((.((..---...))))))))))).....)))-).......------ ( -24.10, z-score = -2.95, R) >droSec1.super_5 2308388 91 - 5866729 CCAACAAUGCACGCAUGCAAAUGUGUGUAGUUCCAUUUCCAGUUCCAGUUUCGUGCAAGCGGCGGCUGCAGCGGAAAAAGGUA-GAAAAAAC------ ((.....(((((((((....)))))))))......(((((.(((.(((((((((....)))).))))).))))))))..))..-........------ ( -26.70, z-score = -1.48, R) >droSim1.chr2L 4153827 91 - 22036055 CCAACAAUGCACGCAUGCAAAUGUGUGUAGUUCCAUUUCCAGUUCCAGUUUCGUGCAAGCGGCGGCUGCAGCGGAAAAAGGUA-GAAGAAAC------ ((.....(((((((((....)))))))))......(((((.(((.(((((((((....)))).))))).))))))))..))..-........------ ( -26.70, z-score = -1.27, R) >droVir3.scaffold_13246 294393 82 - 2672878 CCAGCAAUGUACAGAUGCAAAUGUGUAUA------CGAGUAGUUCCAGUUCUGCGCA----GCUUCUUGGACAUAUUACGGUCAGAGUGCUG------ .(((((.((((((.((....)).))))))------((((.(((((((....)).).)----))).))))(((........)))....)))))------ ( -19.70, z-score = 0.20, R) >droGri2.scaffold_15252 204049 82 + 17193109 CCAGCAAUGCACGCAUGCAAAUGUGUAUA------CGAGUAGUUCCAGUUCUGCGCA----UUGCUUCCAGCAAGUUGCUGCCUGCUUGUUG------ ..((((((((((((((....))))))...------...((((........)))))))----)))))..((((((((........))))))))------ ( -29.70, z-score = -2.38, R) >droWil1.scaffold_180772 1903015 71 - 8906247 CCAAAAAUGUGCUCAUGCAAAUGUCUA------------UAGUUCCAGUUUCUC-------UCUCCUUGCAUCCAAGGAGCUUAACAUAC-------- .........(((....)))........------------........(((....-------.(((((((....)))))))...)))....-------- ( -12.10, z-score = -1.55, R) >consensus CCAACAAUGCACGCAUGCAAAUGUGUGUA_______UUCCAGUUCCAGUUUCGUGCA____GCAGCUGCAGCGGAAAAAGGUA_GAAAAAAC______ .......(((((((((....)))))))))..................................................................... ( -7.52 = -7.63 + 0.11)


| Location | 4,211,233 – 4,211,335 |
|---|---|
| Length | 102 |
| Sequences | 11 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 66.47 |
| Shannon entropy | 0.71539 |
| G+C content | 0.47810 |
| Mean single sequence MFE | -27.81 |
| Consensus MFE | -12.88 |
| Energy contribution | -13.05 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.44 |
| Mean z-score | -0.76 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.744918 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
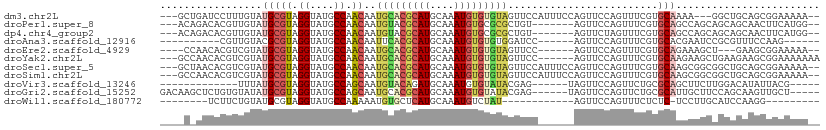
>dm3.chr2L 4211233 102 - 23011544 ---GCUGAUCCUUUGUAUGCGUAGGUAUGCCAACAAUGCACGCAUGCAAAUGUGUGUAGUUCCAUUUCCAGUUCCAGUUUCGUGCAAAA---GGCUGCAGCGGAAAAA-- ---((((..((((((((((.((.((....)).))..(((((((((....)))))))))......................))))).)))---))...)))).......-- ( -30.10, z-score = -1.26, R) >droPer1.super_8 274686 98 + 3966273 ---ACAGACACGUUGUAUGCGUAGGUAUGCCAACAAUGUACGCAUGCAAAUGUGCGCGCUGU-------AGUUCCAGUUUCGUGCAGCCAGCAGCAGCAACUUCAUGG-- ---...(.(((((((((((((((.((.((....)))).))))))))).)))))))(((((((-------.(((.((......)).)))..))))).))..........-- ( -30.40, z-score = 0.24, R) >dp4.chr4_group2 197049 98 - 1235136 ---ACAGACACGUUGUAUGCGUAGGUAUGCCAACAAUGUACGCAUGCAAAUGUGCGCGCUGU-------AGUUCUAGUUUCGUGCAGCCAGCAGCAGCAACUUCAUGG-- ---...(.(((((((((((((((.((.((....)))).))))))))).)))))))..(((((-------.(((((.((.....))))..))).)))))..........-- ( -29.20, z-score = 0.73, R) >droAna3.scaffold_12916 14632977 88 + 16180835 ----------CGUUGUACGCGUAGGUAUGCCAACAAUUCACGCAUGCAAAUGUGUGUGGAUCC------AGUUCCAGUUUCGUGCACGAAUCCGCGUUUCCAAG------ ----------..(((.(((((..((((((..(((....(((((((....)))))))((((...------...))))))).))))).).....)))))...))).------ ( -23.70, z-score = -0.87, R) >droEre2.scaffold_4929 4284581 95 - 26641161 ----CCAACACGUCGUAUGCGUAGGUAUGCCAACAAUGCACGCAUGCAAAUGUGUGUAGUUCC------AGUUCCAGUUUCGUGCAGAAAGCU---GAAGCGGAAAAA-- ----...((((((.((((((((..((((.......))))))))))))..))))))........------..((((.((((((.((.....)))---)))))))))...-- ( -29.30, z-score = -2.01, R) >droYak2.chr2L 4236111 101 - 22324452 ---GCCAACACGUCGUAUGCGUAGGUAUGCCAACAAUGCACGCAUGCAAAUGUGUGUAGUUCC------AGUUCCAGUUUCGUGCAAGAAGCUGAAGAAGCGGAAAAAAA ---....((((((.((((((((..((((.......))))))))))))..))))))....((((------..((((((((((......)))))))..)))..))))..... ( -31.50, z-score = -2.37, R) >droSec1.super_5 2308400 105 - 5866729 ---GCUAACACGUCGUAUGCGUAGGUAUGCCAACAAUGCACGCAUGCAAAUGUGUGUAGUUCCAUUUCCAGUUCCAGUUUCGUGCAAGCGGCGGCUGCAGCGGAAAAA-- ---((((((((((.((((((((..((((.......))))))))))))..)))))).))))....(((((.(((.(((((((((....)))).))))).))))))))..-- ( -34.50, z-score = -1.02, R) >droSim1.chr2L 4153839 105 - 22036055 ---GCCAACACGUCGUAUGCGUAGGUAUGCCAACAAUGCACGCAUGCAAAUGUGUGUAGUUCCAUUUCCAGUUCCAGUUUCGUGCAAGCGGCGGCUGCAGCGGAAAAA-- ---....((((((.((((((((..((((.......))))))))))))..)))))).........(((((.(((.(((((((((....)))).))))).))))))))..-- ( -32.40, z-score = -0.31, R) >droVir3.scaffold_13246 294405 86 - 2672878 -------------UUUAUGCGUAGGUAUGCCAGCAAUGUACAGAUGCAAAUGUGUAUACGAG------UAGUUCCAGUUCUGCGCAGCUUCUUGGACAUAUUACG----- -------------....(((((..((((.........))))..)))))(((((((...((((------.(((((((....)).).)))).)))).)))))))...----- ( -20.80, z-score = -0.01, R) >droGri2.scaffold_15252 204061 99 + 17193109 GACAAGCUCUGUGUAUAUGCGUAGGUAUGCCAGCAAUGCACGCAUGCAAAUGUGUAUACGAG------UAGUUCCAGUUCUGCGCAUUGCUUCCAGCAAGUUGCU----- ...((((..(((((((((((((..((((((((....))...))))))..))))))))))..(------(((........)))))))..))))..(((.....)))----- ( -29.80, z-score = -0.16, R) >droWil1.scaffold_180772 1903027 80 - 8906247 --------UCUUCUGUAUGCGUAGGUAUGCCAAAAAUGUGCUCAUGCAAAUGUCUAU------------AGUUCCAGUUUCUCUC-UCCUUGCAUCCAAGG--------- --------....(((((.((((..(((((.((......))..)))))..)))).)))------------))..............-.(((((....)))))--------- ( -14.20, z-score = -1.34, R) >consensus ____C_AACACGUUGUAUGCGUAGGUAUGCCAACAAUGCACGCAUGCAAAUGUGUGUAGUUCC______AGUUCCAGUUUCGUGCAACCAGCCGCAGAAGCGGAAAAA__ .................(((((.((....)).))..(((((((((....))))))))).........................)))........................ (-12.88 = -13.05 + 0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:15:41 2011