
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 2,803,248 – 2,803,343 |
| Length | 95 |
| Max. P | 0.990063 |

| Location | 2,803,248 – 2,803,343 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 80.09 |
| Shannon entropy | 0.35252 |
| G+C content | 0.41778 |
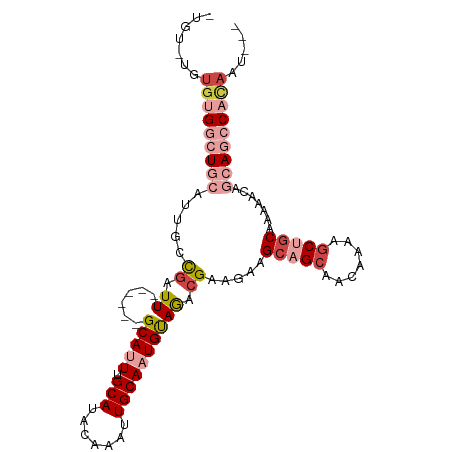
| Mean single sequence MFE | -32.95 |
| Consensus MFE | -19.08 |
| Energy contribution | -21.83 |
| Covariance contribution | 2.75 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.40 |
| SVM RNA-class probability | 0.990063 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
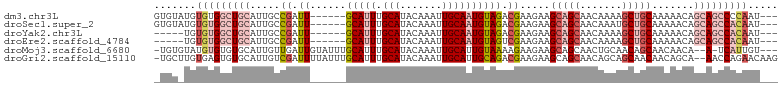
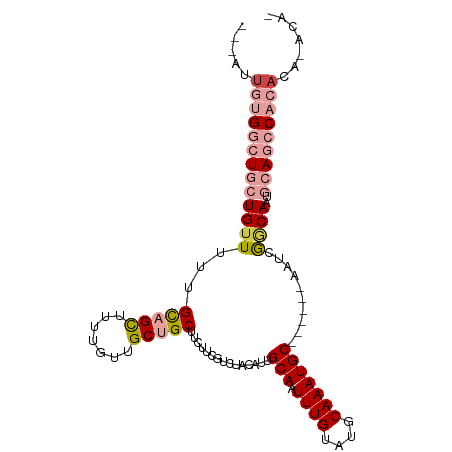
>dm3.chr3L 2803248 95 + 24543557 GUGUAUGUGUGGCUGCAUUGCCGAUU------GCAUUUGCAUACAAAUUGCAAUGUAGACGAAGAAGCAGCAACAAAAGCUGCAAAAACAGCAGCCCCAAU--- .......((.((((((.....((.((------(((((.(((.......)))))))))).)).....(((((.......))))).......)))))).))..--- ( -32.10, z-score = -2.72, R) >droSec1.super_2 2823951 95 + 7591821 GUGUAUGUGUGGCUGCAUUGCCGAUU------GCAUUUGCAUACAAAUUGCAAUGUAGACGAAGAAGCAGCAACAAAUGCUGCAAAAACAGCAGCCACAAU--- .......(((((((((.....((.((------(((((.(((.......)))))))))).)).....((((((.....)))))).......)))))))))..--- ( -38.10, z-score = -4.21, R) >droYak2.chr3L 15066190 90 - 24197627 -----UGUGUGGCUGCAUUGCCGAUU------GCAUUUGCAUACAAAUUGCAAUGUAGACGAAGAAGCAGCAACAAAAGCUGCAAAAACAGCAGCCACAAU--- -----..(((((((((.....((.((------(((((.(((.......)))))))))).)).....(((((.......))))).......)))))))))..--- ( -36.90, z-score = -5.06, R) >droEre2.scaffold_4784 2828582 90 + 25762168 -----UGUGUGGCUGCAUUGCCGAUU------GCAUUUGCAUACAAAUUGCAAUGUAGUCGAAGAAGCAGCAACAAAAGCUGCAAAAACAGCAGCCACAAU--- -----..(((((((((.....(((((------(((((.(((.......))))))))))))).....(((((.......))))).......)))))))))..--- ( -41.80, z-score = -6.51, R) >droMoj3.scaffold_6680 2793415 97 - 24764193 -UGUGUAUGUGUGUGCAUUGUUGAUUGUAUUUGCAUUUGCAUACAAAUUGCAUUGUAAAAGAAGAAGCAGCAACUGCAACAGCAACAACA--A-UCAUUGU--- -(((...(((...((((((((((.((.(.((((((..((((.......)))).))))))).))....)))))).))))...)))...)))--.-.......--- ( -22.90, z-score = 1.18, R) >droGri2.scaffold_15110 19342897 101 + 24565398 -UGCUUGUGAGUGUGCAUUGUCGAUUUUAUUUGCAUUUGCAUACAAAUUGCAUUGCAGACGAAGAAGCAGCAACAGCAGCAACAACAGCA--AACCAGAACAAG -(((((.(..((.((((.((.((((((....(((....)))...)))))))).)))).))..).)))))((....)).((.......)).--............ ( -25.90, z-score = -0.58, R) >consensus _UGU_UGUGUGGCUGCAUUGCCGAUU______GCAUUUGCAUACAAAUUGCAAUGUAGACGAAGAAGCAGCAACAAAAGCUGCAAAAACAGCAGCCACAAU___ .......(((((((((................(((((.(((.......))))))))..........(((((.......))))).......)))))))))..... (-19.08 = -21.83 + 2.75)
| Location | 2,803,248 – 2,803,343 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 80.09 |
| Shannon entropy | 0.35252 |
| G+C content | 0.41778 |
| Mean single sequence MFE | -31.75 |
| Consensus MFE | -18.26 |
| Energy contribution | -20.40 |
| Covariance contribution | 2.14 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.67 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.88 |
| SVM RNA-class probability | 0.972771 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
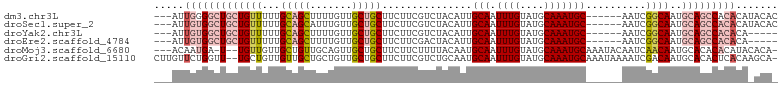
>dm3.chr3L 2803248 95 - 24543557 ---AUUGGGGCUGCUGUUUUUGCAGCUUUUGUUGCUGCUUCUUCGUCUACAUUGCAAUUUGUAUGCAAAUGC------AAUCGGCAAUGCAGCCACACAUACAC ---..((.((((((.......(((((.......)))))............(((((...((((((....))))------))...))))))))))).))....... ( -31.60, z-score = -2.29, R) >droSec1.super_2 2823951 95 - 7591821 ---AUUGUGGCUGCUGUUUUUGCAGCAUUUGUUGCUGCUUCUUCGUCUACAUUGCAAUUUGUAUGCAAAUGC------AAUCGGCAAUGCAGCCACACAUACAC ---..(((((((((.......((((((.....))))))............(((((...((((((....))))------))...))))))))))))))....... ( -37.80, z-score = -4.29, R) >droYak2.chr3L 15066190 90 + 24197627 ---AUUGUGGCUGCUGUUUUUGCAGCUUUUGUUGCUGCUUCUUCGUCUACAUUGCAAUUUGUAUGCAAAUGC------AAUCGGCAAUGCAGCCACACA----- ---..(((((((((.......(((((.......)))))............(((((...((((((....))))------))...))))))))))))))..----- ( -36.30, z-score = -4.27, R) >droEre2.scaffold_4784 2828582 90 - 25762168 ---AUUGUGGCUGCUGUUUUUGCAGCUUUUGUUGCUGCUUCUUCGACUACAUUGCAAUUUGUAUGCAAAUGC------AAUCGGCAAUGCAGCCACACA----- ---..(((((((((.......(((((.......)))))............(((((...((((((....))))------))...))))))))))))))..----- ( -36.30, z-score = -4.19, R) >droMoj3.scaffold_6680 2793415 97 + 24764193 ---ACAAUGA-U--UGUUGUUGCUGUUGCAGUUGCUGCUUCUUCUUUUACAAUGCAAUUUGUAUGCAAAUGCAAAUACAAUCAACAAUGCACACACAUACACA- ---....(((-(--((((((((...(((((.((((............(((((......))))).)))).)))))...))).))))))).))............- ( -20.42, z-score = 0.40, R) >droGri2.scaffold_15110 19342897 101 - 24565398 CUUGUUCUGGUU--UGCUGUUGUUGCUGCUGUUGCUGCUUCUUCGUCUGCAAUGCAAUUUGUAUGCAAAUGCAAAUAAAAUCGACAAUGCACACUCACAAGCA- (((((...(((.--(((.(((((((.(((.(((((.((......))..))))))))((((((((....)))))))).....)))))))))).))).)))))..- ( -28.10, z-score = -1.39, R) >consensus ___AUUGUGGCUGCUGUUUUUGCAGCUUUUGUUGCUGCUUCUUCGUCUACAUUGCAAUUUGUAUGCAAAUGC______AAUCGGCAAUGCAGCCACACA_ACA_ .....(((((((((((((...(((((.......)))))...............(((.((((....)))))))..........))))..)))))))))....... (-18.26 = -20.40 + 2.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:58:32 2011