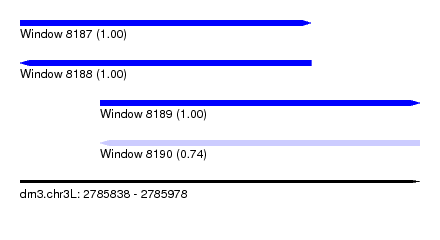
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 2,785,838 – 2,785,978 |
| Length | 140 |
| Max. P | 0.999610 |

| Location | 2,785,838 – 2,785,940 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 65.42 |
| Shannon entropy | 0.66024 |
| G+C content | 0.33685 |
| Mean single sequence MFE | -23.05 |
| Consensus MFE | -14.01 |
| Energy contribution | -14.27 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.56 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.08 |
| SVM RNA-class probability | 0.999610 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
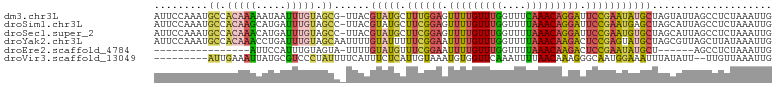
>dm3.chr3L 2785838 102 + 24543557 AUUCCAAAUGCCACAAAAAUAAUUUGUAGCG-UUACGUAUGCUUUGGAGUUUUGUUUGGUUUCAAACAGGAUUCCGAAUAUGCUAGUAUUAGCCUCUAAAUUG ......(((((.(((((.....))))).)))-))..(((((.((((((((((((((((....))))))))))))))))))))).................... ( -29.50, z-score = -4.11, R) >droSim1.chr3L 2339081 102 + 22553184 AUUCCAAAUGCCACAAGCAUGAUUUGUAGCC-UUACGUAUGCUUCGGAGUUUUGUUUGGUUUUAAACAGGAUUCCGAAUGAGCUAGCAUUAGCCUCUAAAUUG .......((((.....))))((((((.((.(-(...((..((((((((((((((((((....)))))))))))))))...)))..))...)).)).)))))). ( -26.20, z-score = -1.95, R) >droSec1.super_2 2806389 102 + 7591821 AUUCCAAAUGCCACAAACAUGAUUUGUAGCC-UUACGUAUGCUUCGGAGUUUUGUUUGGUUUUAAACAGGAUUCCGAAUGUGCUAGCAUUAGCCUCUAAAUUG ......(((((.(((((.....)))))....-....(((((.((((((((((((((((....)))))))))))))))))))))..)))))............. ( -26.50, z-score = -2.46, R) >droYak2.chr3L 15047061 103 - 24197627 AUUCCAAAUGCCACAAACCUGAUUUGUAGCAAUUUUGUAUUUUUCGGAAUUUUGUUUGGUUUUAAACAAGACUCCGAGUAUGCUAGCGUUAGCUUAUAAAUUG ........(((.(((((.....))))).))).....((((..((((((.(((((((((....))))))))).)))))).)))).(((....)))......... ( -24.30, z-score = -2.65, R) >droEre2.scaffold_4784 2811033 80 + 25762168 ----------------AUUCCAUUUGUAGUA-UUUUGUAUGUUUCGGAAUUUUGUUUGGUUUUAAACAAGACUCCGAAUAUGCU------AGCCUCUAAAUUG ----------------..........(((..-....(((((((.((((.(((((((((....))))))))).))))))))))).------.....)))..... ( -19.80, z-score = -3.85, R) >droVir3.scaffold_13049 14887168 92 + 25233164 ---------AUUGAAAUUAUGCGUCCCUAUUUUCAUUUCUCAUUGUAAAUGUGGUUCAAAUUUUAACAAAGGGCAAUGGAAAUUUAUAUU--UUGUUAAAUUG ---------.............(..(((((((.((........)).))))).))..)....((((((((((...(((....)))....))--))))))))... ( -12.00, z-score = 0.35, R) >consensus AUUCCAAAUGCCACAAACAUGAUUUGUAGCA_UUACGUAUGCUUCGGAAUUUUGUUUGGUUUUAAACAAGAUUCCGAAUAUGCUAGCAUUAGCCUCUAAAUUG .........((.(((((.....))))).))......(((((.((((((.(((((((((....))))))))).))))))))))).................... (-14.01 = -14.27 + 0.26)
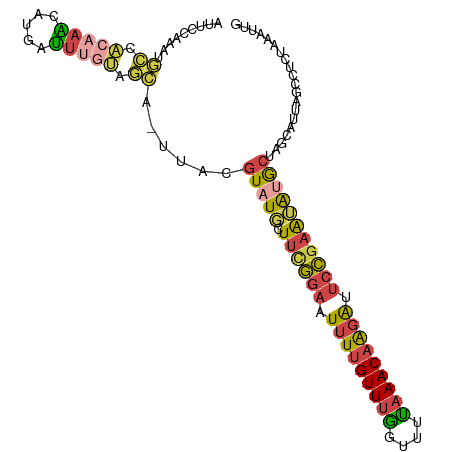
| Location | 2,785,838 – 2,785,940 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 65.42 |
| Shannon entropy | 0.66024 |
| G+C content | 0.33685 |
| Mean single sequence MFE | -19.87 |
| Consensus MFE | -10.53 |
| Energy contribution | -12.38 |
| Covariance contribution | 1.85 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.16 |
| SVM RNA-class probability | 0.997718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 2785838 102 - 24543557 CAAUUUAGAGGCUAAUACUAGCAUAUUCGGAAUCCUGUUUGAAACCAAACAAAACUCCAAAGCAUACGUAA-CGCUACAAAUUAUUUUUGUGGCAUUUGGAAU ...((..((.((((....))))...))..))....((((((....))))))....((((((((....))..-.((((((((.....)))))))).)))))).. ( -23.70, z-score = -2.93, R) >droSim1.chr3L 2339081 102 - 22553184 CAAUUUAGAGGCUAAUGCUAGCUCAUUCGGAAUCCUGUUUAAAACCAAACAAAACUCCGAAGCAUACGUAA-GGCUACAAAUCAUGCUUGUGGCAUUUGGAAU ...(((((((((....))).(((...(((((....(((((......)))))....))))))))........-.(((((((.......))))))).)))))).. ( -22.30, z-score = -1.11, R) >droSec1.super_2 2806389 102 - 7591821 CAAUUUAGAGGCUAAUGCUAGCACAUUCGGAAUCCUGUUUAAAACCAAACAAAACUCCGAAGCAUACGUAA-GGCUACAAAUCAUGUUUGUGGCAUUUGGAAU ...(((((((((....))).((...((((((....(((((......)))))....))))))))........-.(((((((((...))))))))).)))))).. ( -23.10, z-score = -1.55, R) >droYak2.chr3L 15047061 103 + 24197627 CAAUUUAUAAGCUAACGCUAGCAUACUCGGAGUCUUGUUUAAAACCAAACAAAAUUCCGAAAAAUACAAAAUUGCUACAAAUCAGGUUUGUGGCAUUUGGAAU .........(((....))).......(((((((.((((((......)))))).)))))))......((((..((((((((((...)))))))))))))).... ( -28.70, z-score = -4.94, R) >droEre2.scaffold_4784 2811033 80 - 25762168 CAAUUUAGAGGCU------AGCAUAUUCGGAGUCUUGUUUAAAACCAAACAAAAUUCCGAAACAUACAAAA-UACUACAAAUGGAAU---------------- .............------......((((((((.((((((......)))))).))))))))..........-...............---------------- ( -14.20, z-score = -2.81, R) >droVir3.scaffold_13049 14887168 92 - 25233164 CAAUUUAACAA--AAUAUAAAUUUCCAUUGCCCUUUGUUAAAAUUUGAACCACAUUUACAAUGAGAAAUGAAAAUAGGGACGCAUAAUUUCAAU--------- ...........--.......((((((((((.....(((.............)))....))))).)))))((((.((........)).))))...--------- ( -7.22, z-score = 1.04, R) >consensus CAAUUUAGAGGCUAAUGCUAGCAUAUUCGGAAUCCUGUUUAAAACCAAACAAAACUCCGAAGCAUACAUAA_GGCUACAAAUCAUGUUUGUGGCAUUUGGAAU ..........((((....))))...((((((....(((((......)))))....))))))............((((((((.....))))))))......... (-10.53 = -12.38 + 1.85)
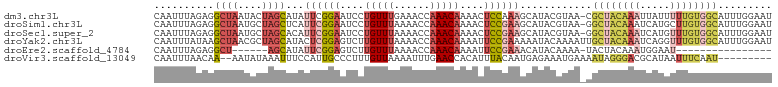
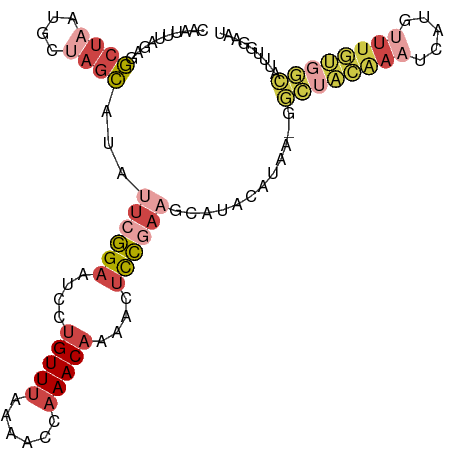
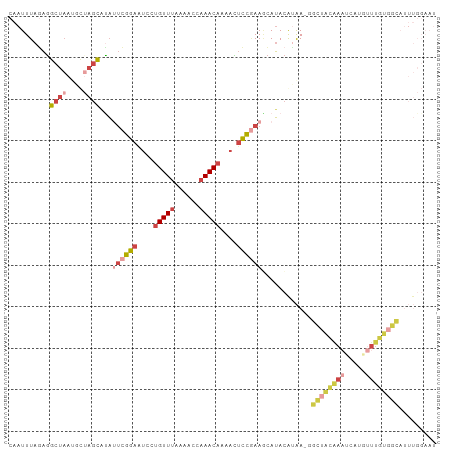
| Location | 2,785,866 – 2,785,978 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 82.22 |
| Shannon entropy | 0.31701 |
| G+C content | 0.37080 |
| Mean single sequence MFE | -28.54 |
| Consensus MFE | -20.58 |
| Energy contribution | -21.62 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.19 |
| Mean z-score | -3.01 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.79 |
| SVM RNA-class probability | 0.995314 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
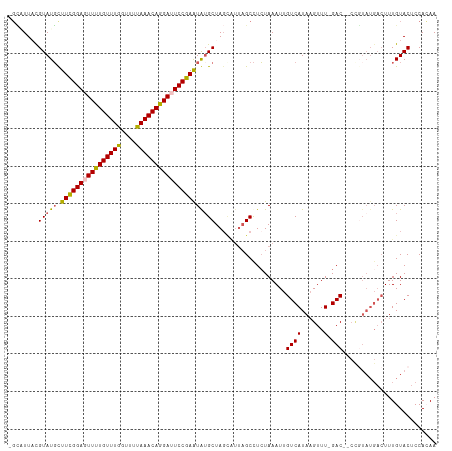
>dm3.chr3L 2785866 112 + 24543557 -GCGUUACGUAUGCUUUGGAGUUUUGUUUGGUUUCAAACAGGAUUCCGAAUAUGCUAGUAUUAGCCUCUAAAUUGUCAUAAGUUU-GACAACCGUAUGACUUUGUACUCCACAA -((..((((((((.((((((((((((((((....)))))))))))))))))))))..)))...)).......((((((......)-)))))..((((......))))....... ( -31.80, z-score = -3.28, R) >droSim1.chr3L 2339109 112 + 22553184 -GCCUUACGUAUGCUUCGGAGUUUUGUUUGGUUUUAAACAGGAUUCCGAAUGAGCUAGCAUUAGCCUCUAAAUUGUCAUAAGUUU-GACCCCCGUAUGACUUUGUACUCCACAA -......((((((.((((((((((((((((....)))))))))))))))).((((((....))).)))......((((......)-)))...))))))................ ( -27.80, z-score = -2.14, R) >droSec1.super_2 2806417 101 + 7591821 -GCCUUACGUAUGCUUCGGAGUUUUGUUUGGUUUUAAACAGGAUUCCGAAUGUGCUAGCAUUAGCCUCUAAAUUGUCAUAAGUUU-GAC-----------UUUGUACACCACAA -.......((((((((((((((((((((((....)))))))))))))))).))((((....)))).........((((......)-)))-----------...))))....... ( -26.20, z-score = -2.21, R) >droYak2.chr3L 15047089 112 - 24197627 GCAAUUUUGUAUUUUUCGGAAUUUUGUUUGGUUUUAAACAAGACUCCGAGUAUGCUAGCGUUAGCUUAUAAAUUGUCAUAAGUUUCGAC--CCUUAUGACUUUGUACUCCAGAA ........((((..((((((.(((((((((....))))))))).)))))).)))).(((....)))((((((..((((((((.......--.))))))))))))))........ ( -28.60, z-score = -3.33, R) >droEre2.scaffold_4784 2811045 105 + 25762168 -GUAUUUUGUAUGUUUCGGAAUUUUGUUUGGUUUUAAACAAGACUCCGAAUAUGCUA------GCCUCUAAAUUGUCAUACGUUUUGAC--CUGUAUGACUUUGUACUCCAGAA -((((...(((((((.((((.(((((((((....))))))))).)))))))))))..------...........((((((((.......--.))))))))...))))....... ( -28.30, z-score = -4.09, R) >consensus _GCAUUACGUAUGCUUCGGAGUUUUGUUUGGUUUUAAACAGGAUUCCGAAUAUGCUAGCAUUAGCCUCUAAAUUGUCAUAAGUUU_GAC__CCGUAUGACUUUGUACUCCACAA ........(((((.((((((((((((((((....)))))))))))))))))))))...................((((((..............)))))).............. (-20.58 = -21.62 + 1.04)

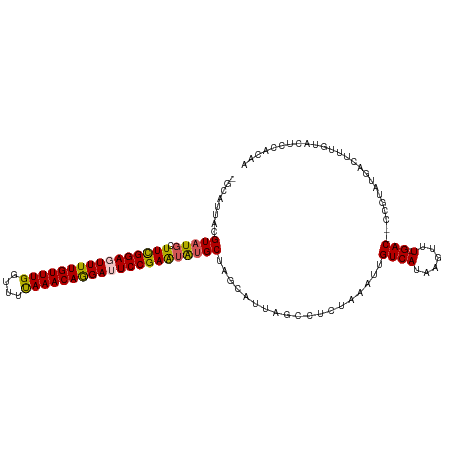
| Location | 2,785,866 – 2,785,978 |
|---|---|
| Length | 112 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 82.22 |
| Shannon entropy | 0.31701 |
| G+C content | 0.37080 |
| Mean single sequence MFE | -25.38 |
| Consensus MFE | -11.62 |
| Energy contribution | -13.62 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.741559 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
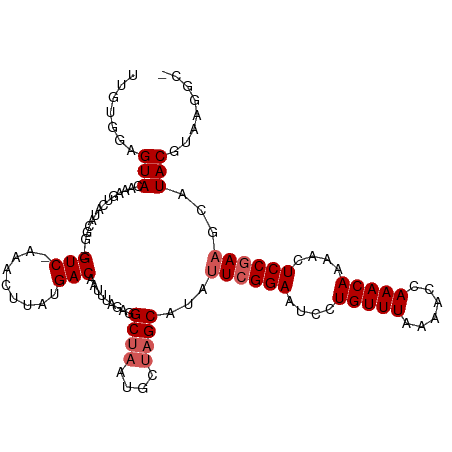
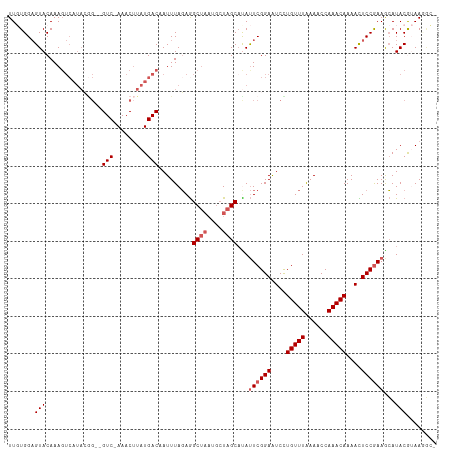
>dm3.chr3L 2785866 112 - 24543557 UUGUGGAGUACAAAGUCAUACGGUUGUC-AAACUUAUGACAAUUUAGAGGCUAAUACUAGCAUAUUCGGAAUCCUGUUUGAAACCAAACAAAACUCCAAAGCAUACGUAACGC- ...((((((.....((((((.((((...-.))))))))))..((..((.((((....))))...))..))....((((((....))))))..))))))...............- ( -25.60, z-score = -2.12, R) >droSim1.chr3L 2339109 112 - 22553184 UUGUGGAGUACAAAGUCAUACGGGGGUC-AAACUUAUGACAAUUUAGAGGCUAAUGCUAGCUCAUUCGGAAUCCUGUUUAAAACCAAACAAAACUCCGAAGCAUACGUAAGGC- .((((((((.....((((((.((.....-...))))))))........(((....))).)))).((((((....(((((......)))))....)))))).))))(....)..- ( -25.00, z-score = -0.95, R) >droSec1.super_2 2806417 101 - 7591821 UUGUGGUGUACAAA-----------GUC-AAACUUAUGACAAUUUAGAGGCUAAUGCUAGCACAUUCGGAAUCCUGUUUAAAACCAAACAAAACUCCGAAGCAUACGUAAGGC- .((((((((.....-----------(((-(......))))........(((....))).)))).((((((....(((((......)))))....)))))).))))(....)..- ( -21.60, z-score = -1.28, R) >droYak2.chr3L 15047089 112 + 24197627 UUCUGGAGUACAAAGUCAUAAGG--GUCGAAACUUAUGACAAUUUAUAAGCUAACGCUAGCAUACUCGGAGUCUUGUUUAAAACCAAACAAAAUUCCGAAAAAUACAAAAUUGC (((..(((((....((((((((.--.......))))))))........(((....)))....)))))(((((.((((((......)))))).)))))))).............. ( -30.40, z-score = -5.03, R) >droEre2.scaffold_4784 2811045 105 - 25762168 UUCUGGAGUACAAAGUCAUACAG--GUCAAAACGUAUGACAAUUUAGAGGC------UAGCAUAUUCGGAGUCUUGUUUAAAACCAAACAAAAUUCCGAAACAUACAAAAUAC- (((((((.......(((((((.(--.......))))))))..)))))))..------.......((((((((.((((((......)))))).)))))))).............- ( -24.30, z-score = -3.75, R) >consensus UUGUGGAGUACAAAGUCAUACGG__GUC_AAACUUAUGACAAUUUAGAGGCUAAUGCUAGCAUAUUCGGAAUCCUGUUUAAAACCAAACAAAACUCCGAAGCAUACGUAAGGC_ .......(((....((((((..............)))))).........((((....))))...((((((....(((((......)))))....))))))...)))........ (-11.62 = -13.62 + 2.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:58:27 2011