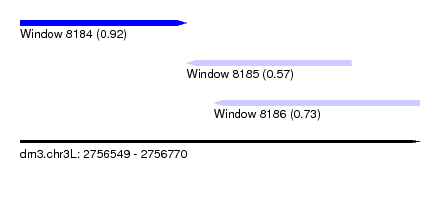
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 2,756,549 – 2,756,770 |
| Length | 221 |
| Max. P | 0.916935 |

| Location | 2,756,549 – 2,756,641 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 71.08 |
| Shannon entropy | 0.50211 |
| G+C content | 0.49008 |
| Mean single sequence MFE | -27.88 |
| Consensus MFE | -11.54 |
| Energy contribution | -12.60 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.25 |
| SVM RNA-class probability | 0.916935 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 2756549 92 + 24543557 AUUUCUUGACACUUCUCUAAAUAGUUUUGCGUGCGACGCAGCAGAAUCGGCGACCAGGGAAAACCAAGGAAAAUGCGAGGAAACUCGCAGGG--------- .(((((((....((((((.....(((((((.(((...)))))))))).((...))))))))...)))))))..((((((....))))))...--------- ( -31.00, z-score = -2.98, R) >droSim1.chr3L 2307380 79 + 22553184 AUUUCUUGACACUUCUCUAAAUAGUUUUGCGUGCUACGCAGCAG-------------GGAAAACCAAGGAAAAUGCGAGGAAACUCGCAGGG--------- .(((((((....((((((.....((..((((.....))))))))-------------))))...)))))))..((((((....))))))...--------- ( -26.20, z-score = -3.31, R) >droSec1.super_2 2776818 79 + 7591821 AUUUCUUGACACUUCUCUAAAUAGUUUUGCGUGCGACGCAGCAG-------------GGAAAACCAAGGAAAAUGCGAGGAAACUCGCAGGG--------- .(((((((....((((((.....((..((((.....))))))))-------------))))...)))))))..((((((....))))))...--------- ( -26.20, z-score = -3.08, R) >droYak2.chr3L 15017588 86 - 24197627 AUUUCUUGACACUUCUCUAAAUAGUUUUGCGUGCGACGCAGCAG------CAGCCAGGGAAAACCAAGGAAAAUGCGAGGAAACUCGCAGGG--------- .(((((((....((((((.....((((((((.....))))).))------)....))))))...)))))))..((((((....))))))...--------- ( -29.60, z-score = -2.87, R) >droVir3.scaffold_13049 14155056 98 - 25233164 AUUUCUCGACACUUCUCUAAAUAGUUUUGCGUGCGACGCAAUGCGAGAGCAAGU---GCAAACUGUGGCAAGAAGGGGGCCAGGUUGGGGAGCACACAGAG .(((((((((.(((((....(((((((((((.....)))).(((....)))...---..)))))))....)))))(....)..)))))))))......... ( -28.20, z-score = -0.24, R) >droMoj3.scaffold_6680 9750211 81 + 24764193 AUUUCUCGACACUUCUCUAAAUAGUUUUGCGUGCGACGCAAUGCGAGAGCAUG----GCAA---GAGCCAAGGGGCGAG--AGGCAAGAG----------- ....(((....((((((.......((((((((((..(((...)))...)))).----))))---))(((....))))))--)))...)))----------- ( -26.10, z-score = -1.35, R) >consensus AUUUCUUGACACUUCUCUAAAUAGUUUUGCGUGCGACGCAGCAG________G____GGAAAACCAAGGAAAAUGCGAGGAAACUCGCAGGG_________ .((((((...(((.........))).(((((.....))))).........................)))))).((((((....))))))............ (-11.54 = -12.60 + 1.06)
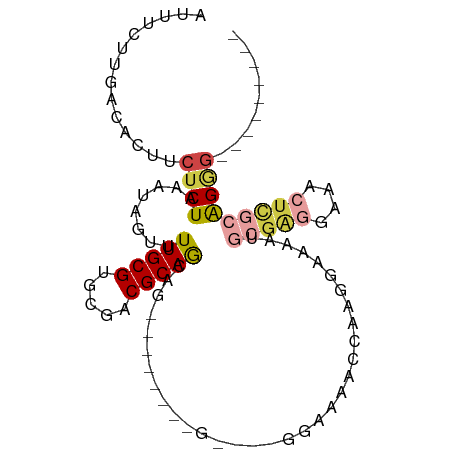
| Location | 2,756,641 – 2,756,732 |
|---|---|
| Length | 91 |
| Sequences | 11 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 77.58 |
| Shannon entropy | 0.49902 |
| G+C content | 0.49973 |
| Mean single sequence MFE | -23.99 |
| Consensus MFE | -17.05 |
| Energy contribution | -16.88 |
| Covariance contribution | -0.17 |
| Combinations/Pair | 1.12 |
| Mean z-score | -0.59 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.567400 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 2756641 91 - 24543557 AAUUUUCGCAACGUUUU-UAAUGGCGUGCGGGGACAUUUUGGCACUCGAGUGACCGUGUGCGUUUUACCUACACCCCAUCCGUGCCGCAUUU- ..((..((((.((((..-....))))))))..))......(((((..(..((...(((((.(.....).)))))..))..))))))......- ( -25.20, z-score = -0.41, R) >droSim1.chr3L 2307459 91 - 22553184 AAUUUUCGCAACGUUUU-UAAUGGCGUGCGGGGACAUUUUGGCACUCGAGUGACCGUGUGCGUUUUACCUACACCCCAACCGUGCCGCAUUU- ..((..((((.((((..-....))))))))..))......(((((..(.((....(((((.(.....).)))))....))))))))......- ( -24.60, z-score = -0.10, R) >droSec1.super_2 2776897 91 - 7591821 AAUUUUCGCAACGUUUU-UAAUGGCGUGCGGGGACAUUUUGGCACUCGAGUGACCGUGUGCGUUUUACCUACACCCCAACCGUGCCGCAUUU- ..((..((((.((((..-....))))))))..))......(((((..(.((....(((((.(.....).)))))....))))))))......- ( -24.60, z-score = -0.10, R) >droYak2.chr3L 15017674 91 + 24197627 AAUUUUCGCAACGUUUU-UAAUGGCGUGCGGGGACAUUUUGGCACUCGAGUGACCGUGUGCGUUUUACCUAUACCCCAUCCACGCCGCAUUU- .................-....((((((.((((.(.....)((((.((......)).))))............))))...))))))......- ( -25.50, z-score = -0.88, R) >droEre2.scaffold_4784 2779399 91 - 25762168 AAUUUUCGAAACGUUUU-UAAUGGCGUGCGGGGACAGUUUGGCACUCGAGUGACCGUGUGCGUUUUACCUACACCCCAACCAUGCCGCUUUU- .................-....((((((.((((........((((.((......)).))))((.......)).))))...))))))......- ( -24.80, z-score = -0.48, R) >droAna3.scaffold_13337 13960653 92 + 23293914 AAUUUUCGCAACAUUUU-UAAUGGCGUGCGGGGACAUUUUGGCACUCGAGUGACCGUGUGCGUUUACCUCCCCUUUCUGCUCCGCAGUCUUUC .......(((.(((...-..)))...)))(((((.......((((.((......)).)))).......)))))...((((...))))...... ( -23.54, z-score = -0.70, R) >droPer1.super_58 136426 91 + 410386 AAUUUUCGCAACAUUUU-UAAUGGCGUGCGGGGACAUUUUGGCACUCGAGUGACCGUAUGCGUGGAUACCCCUGUACCCCUAUACCCCCUAC- .......(((.(((...-..)))...)))((((.......(((((....))).))(((((.(.((.(((....)))))).)))))))))...- ( -22.90, z-score = -0.39, R) >droWil1.scaffold_180916 1628224 86 + 2700594 AAUUUUCGCAACAUUUUCUAAUGGCGUGCGGGGACAUUUUGGCACUCGAGUGACCGUGUGCGU--AUACCAUUCUUCGCUCUCUCUCU----- ..((..((((.((((....))))...))))..)).......((((.((......)).))))..--.......................----- ( -19.30, z-score = -0.33, R) >droVir3.scaffold_13049 14155154 76 + 25233164 AAUUUUCGCAACAUUUU-UAAUGGCGUGCGGGGACAUUUUGGCACUCGAGUGACCGUGUGUCAGUCCGGCCCCCUCU---------------- .......(((.(((...-..)))...)))((((.....(((((((.((......)).))))))).....))))....---------------- ( -23.60, z-score = -0.97, R) >droMoj3.scaffold_6680 9750292 86 - 24764193 AAUUUUCGCAGCAUUUU-UAAUGGCGUGCGGGGACAUUUUGGCACUCGAGUGACCGUGUGUCAGUCCACCUAGGCACA-GUCUCCUGC----- .......(((((((.((-....)).))))((((((.....(((((....))).)).((((((((.....)).))))))-)))))))))----- ( -26.40, z-score = -0.98, R) >droGri2.scaffold_15110 8250916 87 - 24565398 AAUUUUCGCAACAUUUU-UAAUGGCGUACGGGGACAUUUUGGCACUCGAGUGACCGUGUGUCAGUCCCUCUUUUCACACACAGUGUGU----- ......(((..(((...-..))))))...((((((....((((((.((......)).)))))))))))).....(((((...))))).----- ( -23.50, z-score = -1.11, R) >consensus AAUUUUCGCAACAUUUU_UAAUGGCGUGCGGGGACAUUUUGGCACUCGAGUGACCGUGUGCGUUUUACCCACACCCCAACCAUGCCGC_UUU_ ..((..((((.((((....))))...))))..)).......((((.((......)).))))................................ (-17.05 = -16.88 + -0.17)
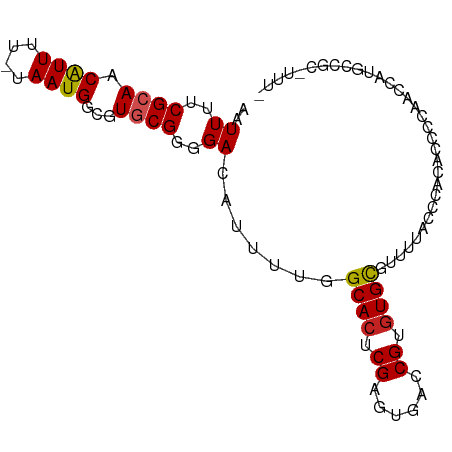
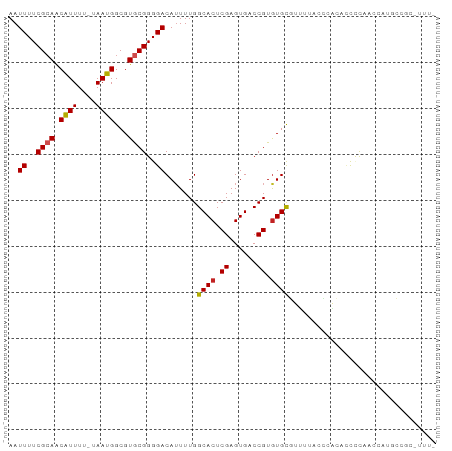
| Location | 2,756,656 – 2,756,770 |
|---|---|
| Length | 114 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 88.84 |
| Shannon entropy | 0.24353 |
| G+C content | 0.49382 |
| Mean single sequence MFE | -33.95 |
| Consensus MFE | -28.32 |
| Energy contribution | -27.74 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.732925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 2756656 114 - 24543557 --GCGCACGCCUCAUUAGAACACCUUUCAGUUUGCUGUGUAAUUUUCGCAACGUUUU-UAAUGGCGUGCGGGGACAUUUUGGCACUCGAGUGACCGUGUGCG---UUUUACCUACACCCC --..(((((((.....(((((......(((....)))(((.......)))..)))))-....)))))))((((........((((.((......)).))))(---(.......)).)))) ( -34.50, z-score = -1.50, R) >droSim1.chr3L 2307474 114 - 22553184 --GCGCACGCCUCAUUAGAACACCUUUCAGUUUGCUGUGUAAUUUUCGCAACGUUUU-UAAUGGCGUGCGGGGACAUUUUGGCACUCGAGUGACCGUGUGCG---UUUUACCUACACCCC --..(((((((.....(((((......(((....)))(((.......)))..)))))-....)))))))((((........((((.((......)).))))(---(.......)).)))) ( -34.50, z-score = -1.50, R) >droSec1.super_2 2776912 114 - 7591821 --GCGCACGCCUCAUUAGAACACCUUUCAGUUUGCUGUGUAAUUUUCGCAACGUUUU-UAAUGGCGUGCGGGGACAUUUUGGCACUCGAGUGACCGUGUGCG---UUUUACCUACACCCC --..(((((((.....(((((......(((....)))(((.......)))..)))))-....)))))))((((........((((.((......)).))))(---(.......)).)))) ( -34.50, z-score = -1.50, R) >droYak2.chr3L 15017689 114 + 24197627 --GCGCACGCCUCAUUAGAACACCUUUCAGUUUGCUGUGUAAUUUUCGCAACGUUUU-UAAUGGCGUGCGGGGACAUUUUGGCACUCGAGUGACCGUGUGCG---UUUUACCUAUACCCC --..(((((((.....(((((......(((....)))(((.......)))..)))))-....)))))))((((.(.....)((((.((......)).)))).---...........)))) ( -33.40, z-score = -1.32, R) >droEre2.scaffold_4784 2779414 114 - 25762168 --GCGCACGCCUCAUUAGAACACCUUUCAGUUUGCUGUGUAAUUUUCGAAACGUUUU-UAAUGGCGUGCGGGGACAGUUUGGCACUCGAGUGACCGUGUGCG---UUUUACCUACACCCC --..(((((((..(((((((.((.((((...((((...)))).....)))).)))))-)))))))))))((((........((((.((......)).))))(---(.......)).)))) ( -33.90, z-score = -1.13, R) >droAna3.scaffold_13337 13960672 111 + 23293914 --GCACACGCCUCAUUAGAACACCUUUCAGUUCGGUGUGUAAUUUUCGCAACAUUUU-UAAUGGCGUGCGGGGACAUUUUGGCACUCGAGUGACCGUGUGCG----UUUACCUCCCCU-- --...((((((......((((........))))(.((((.......)))).).....-....)))))).(((((.......((((.((......)).)))).----......))))).-- ( -33.44, z-score = -1.55, R) >droPer1.super_58 136438 119 + 410386 GGGCGUACGCCUCAUUAGAACACCUUUCAGUUUGCUGUGUAAUUUUCGCAACAUUUU-UAAUGGCGUGCGGGGACAUUUUGGCACUCGAGUGACCGUAUGCGUGGAUACCCCUGUACCCC (((.(((((((......((((........))))(.((((.......)))).).....-....)))))))((((.(((..((((((....))).))).....)))....))))....))). ( -33.10, z-score = -0.05, R) >droWil1.scaffold_180916 1628240 108 + 2700594 --GCGCACGCCUCAUUAGAACACCUUUCAGUUUGCUGUAUAAUUUUCGCAACAUUUUCUAAUGGCGUGCGGGGACAUUUUGGCACUCGAGUGACCGUGUGCGU--AUACCAU-------- --((((((((.(((((.(((.....))).(..(((((.....((..((((.((((....))))...))))..)).....)))))..).)))))..))))))))--.......-------- ( -31.70, z-score = -1.56, R) >droVir3.scaffold_13049 14155163 105 + 25233164 --GCGCACGCCUCAUUAGAACACCUUUCAGUUUGCUGUGUAAUUUUCGCAACAUUUU-UAAUGGCGUGCGGGGACAUUUUGGCACUCGAGUGACCGUGUGUC--AGUCCG---------- --.((((((((......((((........))))(.((((.......)))).).....-....)))))))).((((....((((((.((......)).)))))--))))).---------- ( -35.00, z-score = -2.23, R) >droMoj3.scaffold_6680 9750302 114 - 24764193 --GCGCACGCCUCAUUAGAACACCUUUCAGUUUGCUAUGUAAUUUUCGCAGCAUUUU-UAAUGGCGUGCGGGGACAUUUUGGCACUCGAGUGACCGUGUGUC--AGUCCACCUAGGCAC- --(((((((((......((((........))))(((.((.......)).))).....-....)))))))((((((....((((((.((......)).)))))--))))).))...))..- ( -38.20, z-score = -2.50, R) >droGri2.scaffold_15110 8250927 114 - 24565398 --GCGCACGCCUCAUUAGAACACCUUUCAGUUUGCUGUGUAAUUUUCGCAACAUUUU-UAAUGGCGUACGGGGACAUUUUGGCACUCGAGUGACCGUGUGUC--AGUCCCUCUUUUCAC- --.((.(((((......((((........))))(.((((.......)))).).....-....))))).))(((((....((((((.((......)).)))))--)))))).........- ( -31.20, z-score = -1.54, R) >consensus __GCGCACGCCUCAUUAGAACACCUUUCAGUUUGCUGUGUAAUUUUCGCAACAUUUU_UAAUGGCGUGCGGGGACAUUUUGGCACUCGAGUGACCGUGUGCG___UUUUACCUACACCCC ..(((((((..(((((.((((........))))((((.....((..((((.((((....))))...))))..)).....)))).....))))).)))))))................... (-28.32 = -27.74 + -0.58)
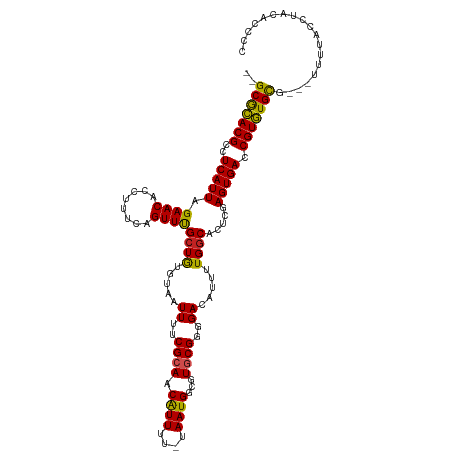
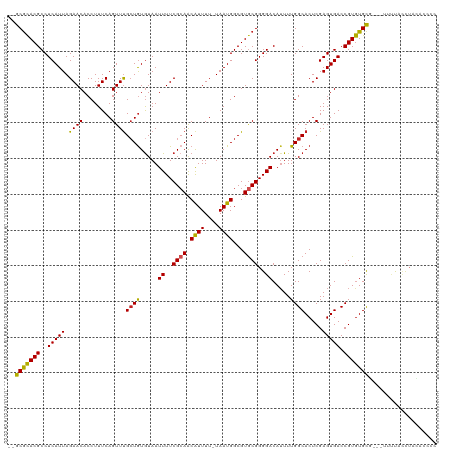
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:58:24 2011