| Sequence ID | dm3.chr3L |
|---|---|
| Location | 2,736,668 – 2,736,719 |
| Length | 51 |
| Max. P | 0.595619 |

| Location | 2,736,668 – 2,736,719 |
|---|---|
| Length | 51 |
| Sequences | 11 |
| Columns | 64 |
| Reading direction | forward |
| Mean pairwise identity | 89.02 |
| Shannon entropy | 0.20477 |
| G+C content | 0.43996 |
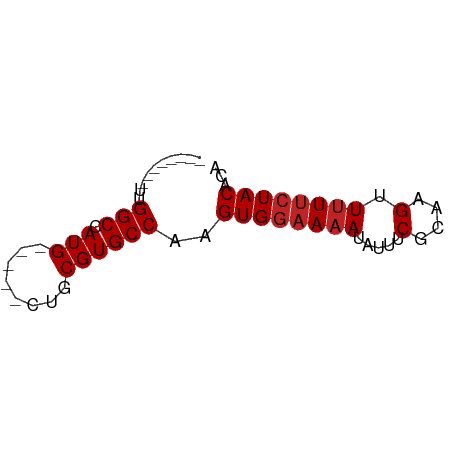
| Mean single sequence MFE | -14.25 |
| Consensus MFE | -11.63 |
| Energy contribution | -11.90 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.00 |
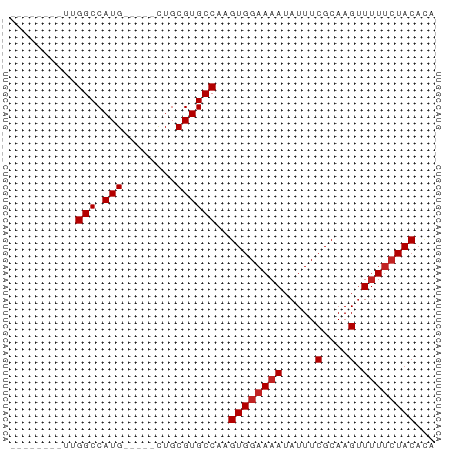
| Mean z-score | -1.50 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.595619 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
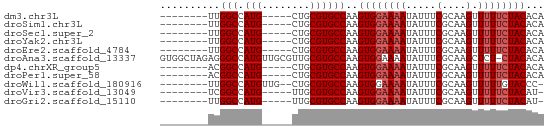
>dm3.chr3L 2736668 51 + 24543557 --------UUGGCCAUG-----CUGCGUGCCAAGUGGAAAAUAUUUCGCAAGUUUUUCUACACA --------(((((.(((-----...))))))))((((((((.....(....).))))))))... ( -14.00, z-score = -1.57, R) >droSim1.chr3L 2292401 51 + 22553184 --------UUGGCCAUG-----CUGCGUGCCAAGUGGAAAAUAUUUCGCAAGUUUUUCUACACA --------(((((.(((-----...))))))))((((((((.....(....).))))))))... ( -14.00, z-score = -1.57, R) >droSec1.super_2 2761656 51 + 7591821 --------UUGGCCAUG-----CUGCGUGCCAAGUGGAAAAUAUUUCGCAAGUUUUUCUACACA --------(((((.(((-----...))))))))((((((((.....(....).))))))))... ( -14.00, z-score = -1.57, R) >droYak2.chr3L 15001535 51 - 24197627 --------UUGGCCAUG-----CUGCGUGCCAAGUGGAAAAUAUUUCGCAAGUUUUUCUACACA --------(((((.(((-----...))))))))((((((((.....(....).))))))))... ( -14.00, z-score = -1.57, R) >droEre2.scaffold_4784 2762809 51 + 25762168 --------UUGGCCAUG-----CUGCGUGCCAAGUGGAAAAUAUUUCGCAAGUUUUUCUACACA --------(((((.(((-----...))))))))((((((((.....(....).))))))))... ( -14.00, z-score = -1.57, R) >droAna3.scaffold_13337 13948469 63 - 23293914 GUGGCUAGAGGGCCAUGUUGCGUUGCGUGCCAAGUGGAAAAUAUUUCGCAAGCUCU-CUACACA (((..(((((((((((((......)))))....((((((....))))))..)))))-)))))). ( -21.70, z-score = -1.69, R) >dp4.chrXR_group5 120307 51 + 740492 --------ACGGCCAUG-----CUGCGUGCCAAGUGGAAAAUAUUUCGCAAGUUUUUCUACACA --------..(((.(((-----...))))))..((((((((.....(....).))))))))... ( -12.70, z-score = -1.03, R) >droPer1.super_58 113229 51 - 410386 --------ACGGCCAUG-----CUGCGUGCCAAGUGGAAAAUAUUUCGCAAGUUUUUCUACACA --------..(((.(((-----...))))))..((((((((.....(....).))))))))... ( -12.70, z-score = -1.03, R) >droWil1.scaffold_180916 1050544 53 + 2700594 --------UUGGCCAUGUUG--CUGCGUGCCAAGUGGAAAAUAUUUCGCAAGUUUUUGUACCC- --------(((((.((((..--..)))))))))((((((....))))))..............- ( -13.00, z-score = -0.97, R) >droVir3.scaffold_13049 14136092 50 - 25233164 --------UCGGCCAUG-----UUGCGUGCCAAGUGGAAAAUAUUUCGCAAGUUUUUCUACAU- --------..(((.(((-----...))))))..((((((((.....(....).))))))))..- ( -12.70, z-score = -1.72, R) >droGri2.scaffold_15110 8233270 50 + 24565398 --------UUGGCCAUG-----UUGCGUGCCAAGUGGAAAAUAUUUCGCAAGUUUUUCUACAU- --------(((((.(((-----...))))))))((((((((.....(....).))))))))..- ( -14.00, z-score = -2.14, R) >consensus ________UUGGCCAUG_____CUGCGUGCCAAGUGGAAAAUAUUUCGCAAGUUUUUCUACACA ..........(((.(((........))))))..((((((((.....(....).))))))))... (-11.63 = -11.90 + 0.27)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:58:21 2011