
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 2,729,387 – 2,729,487 |
| Length | 100 |
| Max. P | 0.997022 |

| Location | 2,729,387 – 2,729,487 |
|---|---|
| Length | 100 |
| Sequences | 13 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 74.44 |
| Shannon entropy | 0.56605 |
| G+C content | 0.49830 |
| Mean single sequence MFE | -26.73 |
| Consensus MFE | -17.77 |
| Energy contribution | -18.27 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.22 |
| Mean z-score | -0.73 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.813990 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
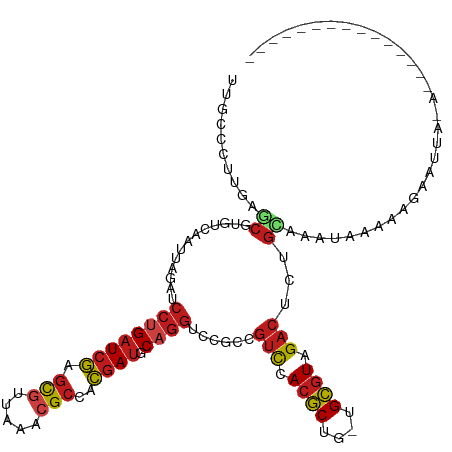
>dm3.chr3L 2729387 100 + 24543557 UUGCCCUUGAGCGUGUCAAUUAGAUCCUGAUCGAGCGUUAAACGCCACGAUGCAGGUCCGCCGUCCACGCUU-UGCGUCGACUCUGCAAAUU---AGAAUUAGA--------------- ..((....(((((((.......((.((((((((.(((.....)))..)))).)))))).......)))))))-.))......((((.....)---)))......--------------- ( -25.74, z-score = -0.41, R) >droSim1.chr3L 2284017 92 + 22553184 UUGCCCUUGAGCGUGUCAAUUAGAUCCUGAUCGAGCGUUAAACGCCACGAUGCAGGUCCGCCGUCCACGCUU-UGCGUCGACUCUGCAAAUUA-------------------------- ((((....(.(((.(((.....)))((((((((.(((.....)))..)))).))))..))))(((.((((..-.)))).)))...))))....-------------------------- ( -25.70, z-score = -0.92, R) >droSec1.super_2 2754774 92 + 7591821 UUGCCCUUGAGCGUGUCAAUUAGAUCCUGAUCGAGCGUUAAAAGCCACGAUGCAGGUCCGCCGUCCACGCUU-UGCGUCGACUCUGCAAAUUA-------------------------- ((((....(.(((.(((.....)))((((((((.((.......))..)))).))))..))))(((.((((..-.)))).)))...))))....-------------------------- ( -23.30, z-score = -0.32, R) >droYak2.chr3L 14994246 109 - 24197627 UUGCCCUUGAGCGUGUCAAUUAGAUCCUGAUCGAGCGUUAAACGCCACGAUGCAGGUCCGCCGUCCACGCUU-UGCGUCGACUCUGUAAAUAUGAAGAAUUAAAAUUAGA--------- ..((....(((((((.......((.((((((((.(((.....)))..)))).)))))).......)))))))-.))..................................--------- ( -25.24, z-score = -0.40, R) >droEre2.scaffold_4784 2755755 109 + 25762168 UUGCCCUUGAGCGUGUCAAUUAGAUCCUGAUCGAGCGUUAAACGCCACGAUGCAGGUCCGCCGUCCACGCUU-UGCGUCGACUCUGCAAAAAUGGAGAAUGGGAAUUAGA--------- ...(((..(((((((.......((.((((((((.(((.....)))..)))).)))))).......)))))))-........((((........))))...))).......--------- ( -31.04, z-score = -0.34, R) >droAna3.scaffold_13337 13941249 103 - 23293914 UUGCCCUUGAGCGUGUCAAAAAGAUCCUGAUCGAGCGUUAGACGCCACGAUGCAGGUCCGCCGUCCACGCUG-UGCGUAGACUCUGCAAUGAAAAAGGAUAAGA--------------- ....((((.((((((.......((.((((((((.(((.....)))..)))).)))))).......)))))).-...((((...)))).......))))......--------------- ( -28.24, z-score = -0.57, R) >dp4.chrXR_group5 111743 100 + 740492 UUACCCUUGAGCGUGUCAAAUAAAUCCUGAUCGAGCGUUAAACGCCACGAUGCAGGUCCUCCGUCCACGCUG-UGCGUAGACUCUGGAAAUAGAUCAAAGA------------------ ......((((.(.............((((((((.(((.....)))..)))).))))(((...(((.((((..-.)))).)))...)))....).))))...------------------ ( -25.40, z-score = -1.16, R) >droPer1.super_58 104534 100 - 410386 UUACCCUUGAGCGUGUCAAAUAAAUCCUGAUCGAGCGUUAAACGCCACGAUGCAGGUCCUCCGUCCACGCUG-UGCGUAGACUCUGGAAAUAGAUCAAAGA------------------ ......((((.(.............((((((((.(((.....)))..)))).))))(((...(((.((((..-.)))).)))...)))....).))))...------------------ ( -25.40, z-score = -1.16, R) >droWil1.scaffold_180916 1041794 102 + 2700594 UUACCCUUUAGCGUGUCUAAUAGAUCCUGAUCGAGCGGUACACGCCACGAUGCUGGGCCGCCAUCCACGCUG-UGCGUAGACUCUG--AAAAAAUAGAAACAAAC-------------- ((((.(..(((((((.......(((....)))(.((((..((.((......))))..)))))...)))))))-.).))))..((((--......)))).......-------------- ( -24.40, z-score = -0.60, R) >droVir3.scaffold_13049 14128534 100 - 25233164 UUUCCCUUUAGCGUAUCAAUAAGAUCCUGAUCGAGCGUUAAACGCCACGAUGCAGGGCCGCCGUCCACGCUG-UGCGUAGACUCUGC-AAUCAGAAAACAUA----------------- ..........((..........(..((((((((.(((.....)))..)))).))))..)...(((.((((..-.)))).)))...))-..............----------------- ( -25.20, z-score = -0.81, R) >droMoj3.scaffold_6680 9723636 110 + 24764193 UUGCCUUUGAGCGUAUCAAUUAGAUCCUGAUCGAGCGUUAAACGCCACGAUGCAGGGCCGCCGUCCACGCUG-UGCGUAGACUCUGCAAAAAAACACGAACAAGAUAGAGA-------- .....((((..(((........(..((((((((.(((.....)))..)))).))))..)((.(((.((((..-.)))).)))...))........)))..)))).......-------- ( -28.40, z-score = -0.49, R) >droGri2.scaffold_15110 8225816 118 + 24565398 UUACCCUUGAGCGUGUCAAUUAGAUCCUGAUCGAGCGUUAAACGCCACGAUGCAGGGCCGCCGUCCACGCUG-UGCGUAGACUCUGUAAGAAAAAUAUGUUUCAAUUAGUGCGUAUACU .........((((((.......(..((((((((.(((.....)))..)))).))))..)......))))))(-((((((....(((...((((......))))...))))))))))... ( -31.52, z-score = -0.95, R) >triCas2.ChLG3 6555594 117 + 32080666 AUAGCGGAGAUGGUAUCAACCAAGUCGCGAUCUAAUUGGAGCUGCCAAAACCCAGAGCC-CGGCUCACUCCGGUGAUUUGACUCUGAAAAAAUAUUUACACUAUUAACAACCCGCAAA- ...((((...((((....))))(((((.((((..(((((((..(((.............-.)))...))))))))))))))))............................))))...- ( -27.94, z-score = -1.42, R) >consensus UUGCCCUUGAGCGUGUCAAUUAGAUCCUGAUCGAGCGUUAAACGCCACGAUGCAGGUCCGCCGUCCACGCUG_UGCGUAGACUCUGCAAAUAAAAAGAAUUA_A_______________ .........................((((((((.(((.....)))..)))).))))......(((.((((....)))).)))..................................... (-17.77 = -18.27 + 0.50)

| Location | 2,729,387 – 2,729,487 |
|---|---|
| Length | 100 |
| Sequences | 13 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 74.44 |
| Shannon entropy | 0.56605 |
| G+C content | 0.49830 |
| Mean single sequence MFE | -33.87 |
| Consensus MFE | -24.13 |
| Energy contribution | -23.86 |
| Covariance contribution | -0.27 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.02 |
| SVM RNA-class probability | 0.997022 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
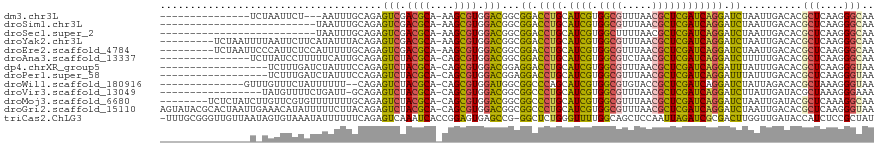
>dm3.chr3L 2729387 100 - 24543557 ---------------UCUAAUUCU---AAUUUGCAGAGUCGACGCA-AAGCGUGGACGGCGGACCUGCAUCGUGGCGUUUAACGCUCGAUCAGGAUCUAAUUGACACGCUCAAGGGCAA ---------------.........---..(((((...(((.((((.-..)))).))).)))))((((.((((.((((.....)))))))))))).............(((....))).. ( -33.40, z-score = -1.95, R) >droSim1.chr3L 2284017 92 - 22553184 --------------------------UAAUUUGCAGAGUCGACGCA-AAGCGUGGACGGCGGACCUGCAUCGUGGCGUUUAACGCUCGAUCAGGAUCUAAUUGACACGCUCAAGGGCAA --------------------------...(((((...(((.((((.-..)))).))).)))))((((.((((.((((.....)))))))))))).............(((....))).. ( -33.40, z-score = -2.31, R) >droSec1.super_2 2754774 92 - 7591821 --------------------------UAAUUUGCAGAGUCGACGCA-AAGCGUGGACGGCGGACCUGCAUCGUGGCUUUUAACGCUCGAUCAGGAUCUAAUUGACACGCUCAAGGGCAA --------------------------...(((((...(((.((((.-..)))).))).)))))((((.((((.(((.......))))))))))).............(((....))).. ( -30.20, z-score = -1.64, R) >droYak2.chr3L 14994246 109 + 24197627 ---------UCUAAUUUUAAUUCUUCAUAUUUACAGAGUCGACGCA-AAGCGUGGACGGCGGACCUGCAUCGUGGCGUUUAACGCUCGAUCAGGAUCUAAUUGACACGCUCAAGGGCAA ---------.......((((((...............(((.((((.-..)))).)))...(((((((.((((.((((.....)))))))))))).)))))))))...(((....))).. ( -31.30, z-score = -1.80, R) >droEre2.scaffold_4784 2755755 109 - 25762168 ---------UCUAAUUCCCAUUCUCCAUUUUUGCAGAGUCGACGCA-AAGCGUGGACGGCGGACCUGCAUCGUGGCGUUUAACGCUCGAUCAGGAUCUAAUUGACACGCUCAAGGGCAA ---------.......(((..(((.((....)).)))(((.((((.-..)))).)))((((..((((.((((.((((.....)))))))))))).((.....))..))))...)))... ( -34.90, z-score = -1.59, R) >droAna3.scaffold_13337 13941249 103 + 23293914 ---------------UCUUAUCCUUUUUCAUUGCAGAGUCUACGCA-CAGCGUGGACGGCGGACCUGCAUCGUGGCGUCUAACGCUCGAUCAGGAUCUUUUUGACACGCUCAAGGGCAA ---------------............(((..((...((((((((.-..)))))))).))(((((((.((((.((((.....)))))))))))).)))...)))...(((....))).. ( -35.30, z-score = -2.34, R) >dp4.chrXR_group5 111743 100 - 740492 ------------------UCUUUGAUCUAUUUCCAGAGUCUACGCA-CAGCGUGGACGGAGGACCUGCAUCGUGGCGUUUAACGCUCGAUCAGGAUUUAUUUGACACGCUCAAGGGUAA ------------------(((((((.(....(((...((((((((.-..))))))))...)))((((.((((.((((.....)))))))))))).............).)))))))... ( -36.00, z-score = -3.27, R) >droPer1.super_58 104534 100 + 410386 ------------------UCUUUGAUCUAUUUCCAGAGUCUACGCA-CAGCGUGGACGGAGGACCUGCAUCGUGGCGUUUAACGCUCGAUCAGGAUUUAUUUGACACGCUCAAGGGUAA ------------------(((((((.(....(((...((((((((.-..))))))))...)))((((.((((.((((.....)))))))))))).............).)))))))... ( -36.00, z-score = -3.27, R) >droWil1.scaffold_180916 1041794 102 - 2700594 --------------GUUUGUUUCUAUUUUUU--CAGAGUCUACGCA-CAGCGUGGAUGGCGGCCCAGCAUCGUGGCGUGUACCGCUCGAUCAGGAUCUAUUAGACACGCUAAAGGGUAA --------------.................--..(.((((((((.-..))))))))..).((((.......((((((((.......(((....)))......))))))))..)))).. ( -26.72, z-score = 0.32, R) >droVir3.scaffold_13049 14128534 100 + 25233164 -----------------UAUGUUUUCUGAUU-GCAGAGUCUACGCA-CAGCGUGGACGGCGGCCCUGCAUCGUGGCGUUUAACGCUCGAUCAGGAUCUUAUUGAUACGCUAAAGGGAAA -----------------....((..((....-.....((((((((.-..))))))))((((..((((.((((.((((.....))))))))))))(((.....))).))))..))..)). ( -36.20, z-score = -2.53, R) >droMoj3.scaffold_6680 9723636 110 - 24764193 --------UCUCUAUCUUGUUCGUGUUUUUUUGCAGAGUCUACGCA-CAGCGUGGACGGCGGCCCUGCAUCGUGGCGUUUAACGCUCGAUCAGGAUCUAAUUGAUACGCUCAAAGGCAA --------........(((..((((((..((.((...((((((((.-..)))))))).))(..((((.((((.((((.....))))))))))))..).))..))))))..)))...... ( -38.30, z-score = -2.38, R) >droGri2.scaffold_15110 8225816 118 - 24565398 AGUAUACGCACUAAUUGAAACAUAUUUUUCUUACAGAGUCUACGCA-CAGCGUGGACGGCGGCCCUGCAUCGUGGCGUUUAACGCUCGAUCAGGAUCUAAUUGACACGCUCAAGGGUAA ......(((.((....((((......))))....)).((((((((.-..)))))))).)))(((((....(((((((.....)))(((((.((...)).)))))))))....))))).. ( -35.10, z-score = -1.87, R) >triCas2.ChLG3 6555594 117 - 32080666 -UUUGCGGGUUGUUAAUAGUGUAAAUAUUUUUUCAGAGUCAAAUCACCGGAGUGAGCCG-GGCUCUGGGUUUUGGCAGCUCCAAUUAGAUCGCGACUUGGUUGAUACCAUCUCCGCUAU -.(((.((((((((((...............(((((((((...((((....))))....-)))))))))..))))))))))))).......(((...((((....))))....)))... ( -33.43, z-score = -0.89, R) >consensus _______________U_UAAUUCUAUUUAUUUGCAGAGUCUACGCA_CAGCGUGGACGGCGGACCUGCAUCGUGGCGUUUAACGCUCGAUCAGGAUCUAAUUGACACGCUCAAGGGCAA ..................................((((((.((((....)))).)))......((((.((((.((((.....)))))))))))).))).........(((....))).. (-24.13 = -23.86 + -0.27)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:58:20 2011