
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 2,690,555 – 2,690,689 |
| Length | 134 |
| Max. P | 0.792038 |

| Location | 2,690,555 – 2,690,689 |
|---|---|
| Length | 134 |
| Sequences | 7 |
| Columns | 159 |
| Reading direction | forward |
| Mean pairwise identity | 66.52 |
| Shannon entropy | 0.61331 |
| G+C content | 0.51106 |
| Mean single sequence MFE | -42.89 |
| Consensus MFE | -17.47 |
| Energy contribution | -19.62 |
| Covariance contribution | 2.15 |
| Combinations/Pair | 1.39 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.792038 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 2690555 134 + 24543557 -----------------UUUUACCAAACUAUUUGGCUGCUGUGCUAAGCUC--UUUGGCUGCCA-GAUAAGCUGCUCUAGAUCGUCGCCAUCUA-GCAGUGCAAUCACAAUGCUUUGGCCAGCUUAUCGAGCGUUCACUGCAGCUGGGCUGCACU---- -----------------.............((..(((((.(((....((((--..(((...)))-(((((((((.....(.....)((((...(-(((.((......)).)))).)))))))))))))))))...))).)))))..)).......---- ( -45.30, z-score = -0.69, R) >droWil1.scaffold_180916 991464 147 + 2700594 UUUUGUUAAGCCCUUCGCUUACUCUUUGCUCUUAAGCCCUGCCAUAAACGCAGUCCAGCCCCACCCAUCCCC-GUCUC---UCACCCCCAUUU--------CAAUCACAAUGCUUUGUACAGCUUAUCGGCCAUUCACUGCAAUUCGCCAGAAUGGAUG ......(((((.....)))))......(((..(((((.((((.......))))...................-.....---............--------.....((((....))))...)))))..)))(((((...((.....))..))))).... ( -21.70, z-score = -0.31, R) >droEre2.scaffold_4784 2711008 135 + 25762168 -----------------UUCCAGCCAACUAUUUGGCUGCUGUCCUAAGCUC--CUUGGCUGCCUAGAUAAGCUGCUAUAGAUCCUCGCCAGCAA-GCAGUGCAAUCACAAUGCUUUGGCCAGCUUAUCGGGCGUUCACUGCAGCUGGGCUGCACU---- -----------------....((((((....))))))((.((((..(((((--..((..(((((.(((((((((.....(.....)((((..((-(((.((......)).)))))))))))))))))))))))..))..).)))))))).))...---- ( -50.40, z-score = -1.10, R) >droYak2.chr3L 14955236 135 - 24197627 -----------------UCCCACCCGACUAUUUGGCUGCUGUCCUAAGCUC--UUUGGCUGUCU-GAUAAGCUGCUCUAGAUCCGCGCCAUCAACGCGGCGCAAUCACAAUGCUUUGGCCAGCUUAUCGGGCGUUCACUGCAGCUGGGCUGCACU---- -----------------......((((....)))).(((.((((..(((((--..((..(((((-((((((((((....(((..(((((........))))).))).(((....)))).))))))))))))))..))..).)))))))).)))..---- ( -49.90, z-score = -1.91, R) >droSec1.super_2 2716268 134 + 7591821 -----------------UUUUACCAAACUAUUUGGCUGCUGUGCUAAGCUC--UUUGGCUGCCU-GAUAAGCUGCUCUAGAUCCGCGCCAUCUA-GUAGUGCAAUCAAAAUGCUUUGGCCAGCUUAUCGGGCGUUCACUGCAGCUGGGCUGCACU---- -----------------.............((..(((((.(((...((((.--...))))((((-(((((((((((((((((.......)))))-)....(((.......)))...)).)))))))))))))...))).)))))..)).......---- ( -46.80, z-score = -1.16, R) >droSim1.chr3L 2246633 134 + 22553184 -----------------UUUUACAAAACUAUUUGGCUGCUGUGCUAAGCUC--UUUGGCUGCCU-GAUAAGCUGCCCUAGAUCCUCGCCAUCAA-GCAGUGCAAUCACAAUGCUUUGGCCAGCUUAUCGGGCGUUCACUGCAGCUGGGCUGCACU---- -----------------.............((..(((((.(((...((((.--...))))((((-(((((((((.....(.....)((((..((-(((.((......)).))))))))))))))))))))))...))).)))))..)).......---- ( -50.10, z-score = -2.04, R) >droVir3.scaffold_13049 14079235 109 - 25233164 ------------------------------UUUCUCUGUUGGCGCAAAUCC---UUGGGUGUACUCGUAA---GCUGC---UCACAGCCAUUCA-------CAACUCUUGCGCUUUGUACAGCUUAUGGUACAUUCACAGCAAGUCUGUUGCCCU---- ------------------------------..........((.((((....---.(((((((((.(((((---((((.---.((.(((((....-------.......)).))).))..))))))))))))))))))(((.....))))))))).---- ( -36.00, z-score = -3.32, R) >consensus _________________UUUUACCAAACUAUUUGGCUGCUGUCCUAAGCUC__UUUGGCUGCCU_GAUAAGCUGCUCUAGAUCCUCGCCAUCAA_GCAGUGCAAUCACAAUGCUUUGGCCAGCUUAUCGGGCGUUCACUGCAGCUGGGCUGCACU____ ..............................((..(((((.......((((......))))((((.(((((((((............((((.........................))))))))))))))))).......)))))..))........... (-17.47 = -19.62 + 2.15)
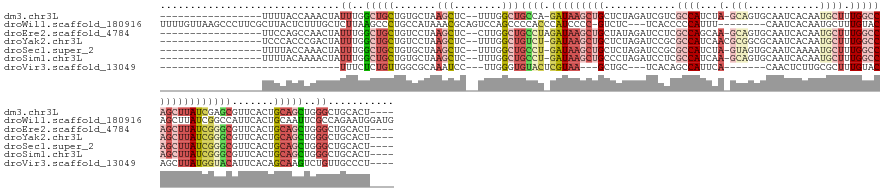

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:58:12 2011