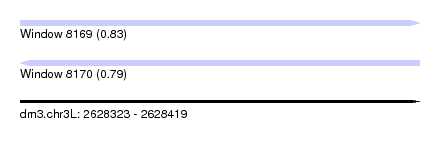
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 2,628,323 – 2,628,419 |
| Length | 96 |
| Max. P | 0.829241 |

| Location | 2,628,323 – 2,628,419 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 89.68 |
| Shannon entropy | 0.16999 |
| G+C content | 0.48106 |
| Mean single sequence MFE | -22.96 |
| Consensus MFE | -18.76 |
| Energy contribution | -19.16 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.83 |
| SVM RNA-class probability | 0.829241 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
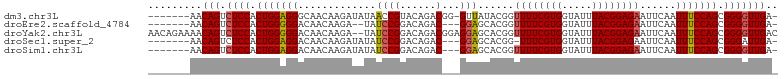
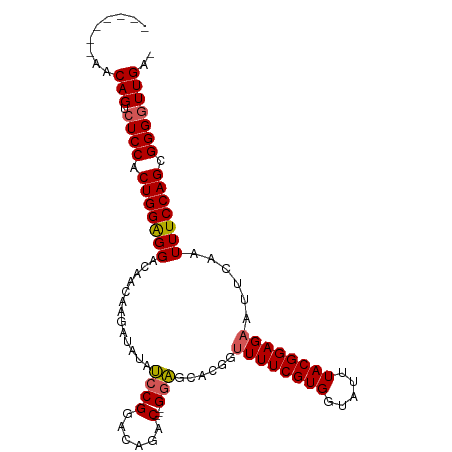
>dm3.chr3L 2628323 96 + 24543557 -UCAACCCCGCUGGAAAUUGAAUUCUCCGUAAAUACCACGAAAACCGUAUAAC-CCGUCUGUACGGUUAUAUCUUGUUGCCCUCCAGUGGAGACUGUU------- -......((((((((............(((.......)))..(((((((((..-.....)))))))))..............))))))))........------- ( -24.50, z-score = -2.76, R) >droEre2.scaffold_4784 2646911 92 + 25762168 -UCAACCCCGCUGGAAAUUGAAUUCUCCGUAAAUACCACGAAAACCGUGCUCC---GUCUGUCCGGAUA--UCUUGUUGUCCCCCAGUGGAGACUGUU------- -......(((((((......................((((.....))))....---........(((((--......))))).)))))))........------- ( -21.10, z-score = -1.44, R) >droYak2.chr3L 14888942 103 - 24197627 GUCAACCCCGCUGGAAAUUGAAUUCUCCGUAAAUACCACGAAAACCGUGCUCCUCCGUCUGUCCGGAUA--UCUUGUUGUCCCCCAGUGGAGACUGUUUUCUGUU (((....(((((((......................((((.....))))...............(((((--......))))).))))))).)))........... ( -22.20, z-score = -1.30, R) >droSec1.super_2 2651828 93 + 7591821 -UCAAUCCCGCUGGAAAUUGAAUUCUCCGUAAAUACCACGAAA-CCGUGCUCC---GUCUGUCCGGAUAUAUCUUGUUGUCCUCCAGUGGAGACUGUU------- -.((.((((((((((.....................((((...-.))))....---........(((((........))))))))))))).)).))..------- ( -24.30, z-score = -2.43, R) >droSim1.chr3L 2183756 94 + 22553184 -UCAACCCCGCUGGAAAUUGAAUUCUCCGUAAAUACCACGAAAACCGUGCUCC---GUCUGUCCGGAUAUAUCUUGUUGUCCUCCAGUGGAGACUGUU------- -......((((((((.....................((((.....))))....---........(((((........)))))))))))))........------- ( -22.70, z-score = -1.95, R) >consensus _UCAACCCCGCUGGAAAUUGAAUUCUCCGUAAAUACCACGAAAACCGUGCUCC___GUCUGUCCGGAUAUAUCUUGUUGUCCUCCAGUGGAGACUGUU_______ .......(((((((......................((((.....))))...............(((((........))))).)))))))............... (-18.76 = -19.16 + 0.40)

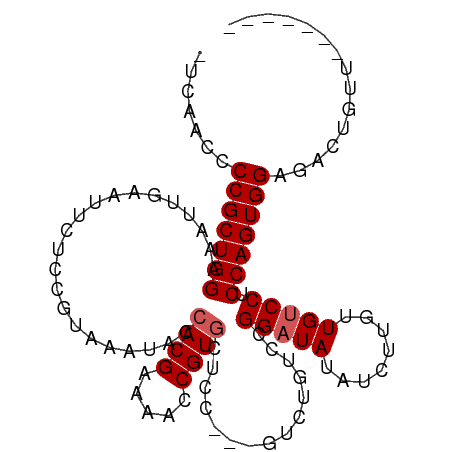
| Location | 2,628,323 – 2,628,419 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 89.68 |
| Shannon entropy | 0.16999 |
| G+C content | 0.48106 |
| Mean single sequence MFE | -28.66 |
| Consensus MFE | -22.21 |
| Energy contribution | -22.37 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.785779 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 2628323 96 - 24543557 -------AACAGUCUCCACUGGAGGGCAACAAGAUAUAACCGUACAGACGG-GUUAUACGGUUUUCGUGGUAUUUACGGAGAAUUCAAUUUCCAGCGGGGUUGA- -------..(((.((((.(((((((....)..(((((((((((....)).)-))))))...((((((((.....))))))))..))...)))))).))))))).- ( -29.10, z-score = -2.41, R) >droEre2.scaffold_4784 2646911 92 - 25762168 -------AACAGUCUCCACUGGGGGACAACAAGA--UAUCCGGACAGAC---GGAGCACGGUUUUCGUGGUAUUUACGGAGAAUUCAAUUUCCAGCGGGGUUGA- -------..(((.((((.((((..(.......(.--..((((......)---))).)....((((((((.....))))))))......)..)))).))))))).- ( -28.30, z-score = -1.86, R) >droYak2.chr3L 14888942 103 + 24197627 AACAGAAAACAGUCUCCACUGGGGGACAACAAGA--UAUCCGGACAGACGGAGGAGCACGGUUUUCGUGGUAUUUACGGAGAAUUCAAUUUCCAGCGGGGUUGAC .........(((.((((.((((..(.........--..((((......))))(((......((((((((.....)))))))).)))..)..)))).))))))).. ( -28.50, z-score = -1.49, R) >droSec1.super_2 2651828 93 - 7591821 -------AACAGUCUCCACUGGAGGACAACAAGAUAUAUCCGGACAGAC---GGAGCACGG-UUUCGUGGUAUUUACGGAGAAUUCAAUUUCCAGCGGGAUUGA- -------..(((((.((.((((((.....)..((....((((.((..((---(((((...)-)))))).)).....))))....))....))))).))))))).- ( -29.20, z-score = -2.76, R) >droSim1.chr3L 2183756 94 - 22553184 -------AACAGUCUCCACUGGAGGACAACAAGAUAUAUCCGGACAGAC---GGAGCACGGUUUUCGUGGUAUUUACGGAGAAUUCAAUUUCCAGCGGGGUUGA- -------..(((.((((.(((((((.............((((......)---)))......((((((((.....))))))))......))))))).))))))).- ( -28.20, z-score = -2.09, R) >consensus _______AACAGUCUCCACUGGAGGACAACAAGAUAUAUCCGGACAGAC___GGAGCACGGUUUUCGUGGUAUUUACGGAGAAUUCAAUUUCCAGCGGGGUUGA_ .........(((.((((.(((((((..............(((................)))((((((((.....))))))))......))))))).))))))).. (-22.21 = -22.37 + 0.16)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:58:11 2011