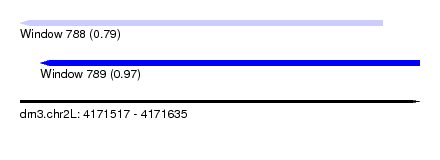
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,171,517 – 4,171,635 |
| Length | 118 |
| Max. P | 0.973304 |

| Location | 4,171,517 – 4,171,624 |
|---|---|
| Length | 107 |
| Sequences | 7 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 78.19 |
| Shannon entropy | 0.39648 |
| G+C content | 0.56272 |
| Mean single sequence MFE | -23.21 |
| Consensus MFE | -11.25 |
| Energy contribution | -11.50 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.794669 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
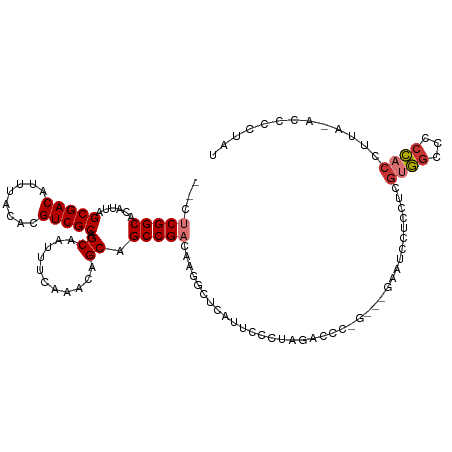
>dm3.chr2L 4171517 107 - 23011544 AGCGACAUUUACACGUCGCAGCAAUUUCAAACAGCAGCCGACAAGGCUCAUUCCCUAGACCCGGGAUGAUUCCUCCGCGUGGCCCCCACCUUAACCCCUAUAGCUCC .(((((........)))))(((...........((.(((.....)))(((((((........))))))).......))((((...)))).............))).. ( -28.50, z-score = -2.61, R) >droEre2.scaffold_4929 4239113 104 - 26641161 AGCGACAUUUACACGUCGCAGCAAUUUCAAACAGCAGCCGACAAGGCUCAUUCCCUAGACCCAG---GAAUCCUCCUCGUGGCCCCCACCUUAACCCCUAUAGCACC .(((((........))))).((...........(.((((.....)))))......(((....((---((....)))).((((...))))........)))..))... ( -22.70, z-score = -2.58, R) >droYak2.chr2L 4198232 104 - 22324452 AGCGACAUUUACACGUCGCAGCAAUUUCAAACAGCAGCCGACAAGGCUCAUUCCCUAGACCCAG---GAAUCCUUCUCGUGGCCCCCACCUUAACCCCUAUAGCACC .(((((........))))).((...........(.((((.....)))))(((((.........)---)))).......((((...)))).............))... ( -21.50, z-score = -2.13, R) >droSec1.super_5 2271098 104 - 5866729 AGCGACAUUUACACGUCGCAGCAAUUUCAAACAGCAGCCGACAAGGCUCAUUCCCUAGACCCGG---GAUUCCUCCGCGUGGCCCCCACCUUAACCCCCAUAGCACC .(((((........))))).((...........((((((.....))))...((((.......))---)).......))((((...)))).............))... ( -26.60, z-score = -2.88, R) >droSim1.chr2L 4127236 104 - 22036055 AGCGACAUUUACACGUCGCAGCAAUUUCAAACAGCAGCCGACAAGGCUCAUUCCCUAGACCCGG---GAUUCCUCCGCGUGGCCCCCACCUUAACCCCCAUAGCACC .(((((........))))).((...........((((((.....))))...((((.......))---)).......))((((...)))).............))... ( -26.60, z-score = -2.88, R) >droPer1.super_1 7073857 87 + 10282868 AGCGACAUUUACACGUCGCAGCAAUUUCAAACAGCAGCCGACA--UCCCAGCCACCGCCUCCUGG-UAGCGCCUCCAUGUAGCUCCUCUC----------------- .(((((........))))).............((.(((..(((--(....((.((((.....)))-).))......)))).))).))...----------------- ( -18.30, z-score = -1.27, R) >dp4.chr4_group3 9954149 87 + 11692001 AGCGACAUUUACACGUCGCAGCAAUUUCAAACAGCAGCCGACA--UCCCAGCCACCGCCUCUUGG-UAGCGCCUCCAUGUAGCUCCUCUC----------------- .(((((........))))).............((.(((..(((--(....((.((((.....)))-).))......)))).))).))...----------------- ( -18.30, z-score = -1.22, R) >consensus AGCGACAUUUACACGUCGCAGCAAUUUCAAACAGCAGCCGACAAGGCUCAUUCCCUAGACCCGG___GAUUCCUCCGCGUGGCCCCCACCUUAACCCCUAUAGCACC .(((((........))))).((...........)).(((.....)))...............................((((...)))).................. (-11.25 = -11.50 + 0.25)
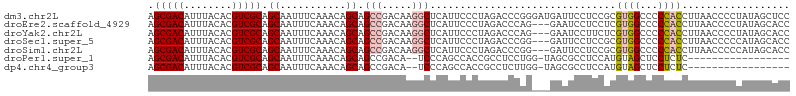
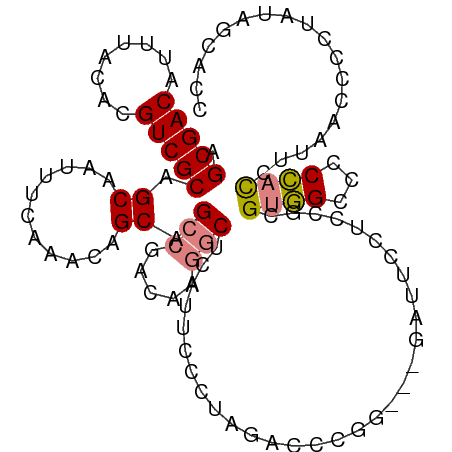
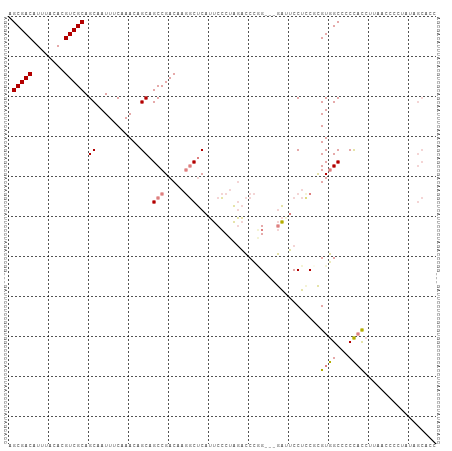
| Location | 4,171,523 – 4,171,635 |
|---|---|
| Length | 112 |
| Sequences | 8 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 79.37 |
| Shannon entropy | 0.38494 |
| G+C content | 0.54936 |
| Mean single sequence MFE | -24.95 |
| Consensus MFE | -17.00 |
| Energy contribution | -17.38 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.89 |
| SVM RNA-class probability | 0.973304 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 4171523 112 - 23011544 --CUCGGCACAUUAGCGACAUUUACACGUCGCAGCAAUUUCAAACAGCAGCCGACAAGGCUCAUUCCCUAGACCCGGGAUGAUUCCUCCGCGUGGCCCCCACCUUA-ACCCCUAU --.(((((......(((((........))))).((...........)).)))))..(((.(((((((........)))))))..)))....((((...))))....-........ ( -29.60, z-score = -2.34, R) >droEre2.scaffold_4929 4239119 109 - 26641161 --CUCGGCACAUUAGCGACAUUUACACGUCGCAGCAAUUUCAAACAGCAGCCGACAAGGCUCAUUCCCUAGACCCAG---GAAUCCUCCUCGUGGCCCCCACCUUA-ACCCCUAU --.(((((......(((((........))))).((...........)).)))))...((.((........)).))((---((....)))).((((...))))....-........ ( -25.30, z-score = -2.59, R) >droYak2.chr2L 4198238 109 - 22324452 --CUCGGCACAUUAGCGACAUUUACACGUCGCAGCAAUUUCAAACAGCAGCCGACAAGGCUCAUUCCCUAGACCCAG---GAAUCCUUCUCGUGGCCCCCACCUUA-ACCCCUAU --.(((((......(((((........))))).((...........)).))))).((((...(((((.........)---))))))))...((((...))))....-........ ( -25.10, z-score = -2.53, R) >droSec1.super_5 2271104 109 - 5866729 --CUCGGCACAUUAGCGACAUUUACACGUCGCAGCAAUUUCAAACAGCAGCCGACAAGGCUCAUUCCCUAGACCCGG---GAUUCCUCCGCGUGGCCCCCACCUUA-ACCCCCAU --...(((.(((..(((((........))))).((...........(.((((.....)))))..((((.......))---)).......)))))))).........-........ ( -27.70, z-score = -2.25, R) >droSim1.chr2L 4127242 109 - 22036055 --CUCGGCACAUUAGCGACAUUUACACGUCGCAGCAAUUUCAAACAGCAGCCGACAAGGCUCAUUCCCUAGACCCGG---GAUUCCUCCGCGUGGCCCCCACCUUA-ACCCCCAU --...(((.(((..(((((........))))).((...........(.((((.....)))))..((((.......))---)).......)))))))).........-........ ( -27.70, z-score = -2.25, R) >droPer1.super_1 7073858 99 + 10282868 CUCGCGGCACAUUAGCGACAUUUACACGUCGCAGCAAUUUCAAACAGCAGCCGACA--UCCCAGCC--------------ACCGCCUCCUGGUAGCGCCUCCAUGUAGCUCCUCU ...(((((......(((((........))))).((...........)).))).(((--(....((.--------------((((.....)))).))......)))).))...... ( -23.30, z-score = -1.22, R) >dp4.chr4_group3 9954150 99 + 11692001 CUCGCGGCACAUUAGCGACAUUUACACGUCGCAGCAAUUUCAAACAGCAGCCGACA--UCCCAGCC--------------ACCGCCUCUUGGUAGCGCCUCCAUGUAGCUCCUCU ...(((((......(((((........))))).((...........)).))).(((--(....((.--------------((((.....)))).))......)))).))...... ( -23.30, z-score = -1.20, R) >droWil1.scaffold_180703 1772167 90 + 3946847 --CUCGGCACAUUAGCGACAUUUACACGUCGCAGCAAUUUCAAACAGCAGCCGACA----UCAU-----------------AAUGCUCCUUCUGCCCCUCACCUAAUACUCCU-- --.(((((......(((((........))))).((...........)).)))))..----....-----------------...((.......))..................-- ( -17.60, z-score = -3.08, R) >consensus __CUCGGCACAUUAGCGACAUUUACACGUCGCAGCAAUUUCAAACAGCAGCCGACAAGGCUCAUUCCCUAGACCC_G___GAAUCCUCCUCGUGGCCCCCACCUUA_ACCCCUAU ...(((((......(((((........))))).((...........)).))))).....................................((((...))))............. (-17.00 = -17.38 + 0.38)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:15:38 2011