| Sequence ID | dm3.chr3L |
|---|---|
| Location | 2,552,630 – 2,552,785 |
| Length | 155 |
| Max. P | 0.993463 |

| Location | 2,552,630 – 2,552,785 |
|---|---|
| Length | 155 |
| Sequences | 4 |
| Columns | 172 |
| Reading direction | forward |
| Mean pairwise identity | 75.36 |
| Shannon entropy | 0.38332 |
| G+C content | 0.28884 |
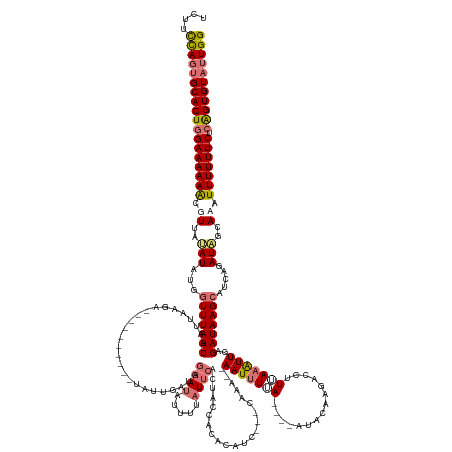
| Mean single sequence MFE | -34.35 |
| Consensus MFE | -15.35 |
| Energy contribution | -17.29 |
| Covariance contribution | 1.94 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.50 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.993463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
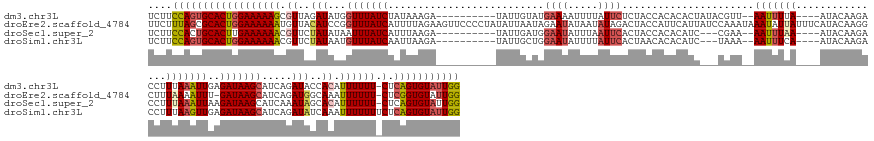
>dm3.chr3L 2552630 155 + 24543557 UCUUCCAGUGCACUGGAAAAAGCGUUAGAUAUGGUUUAUCUAUAAAGA----------UAUUGUAUGAAAAUUUUAUUCUCUACCACACACUAUACGUU--AAUUUUA----AUACAAGACCUUUAAAUUGAGAUAAGCAUCAGAUACCACAUUUUUU-CUCAGUGUAUUGG ....((((((((((((((((((.((...((...((((((((.(((((.----------..((((((.((((((..........................--)))))).----))))))...))))).....)))))))).....))...)).))))))-).))))))))))) ( -37.77, z-score = -4.02, R) >droEre2.scaffold_4784 2568718 170 + 25762168 UUCUUUAGCGCACUGGAAAAAAUGUUACAUCCGGUUUAUCAUUUUAGAAGUUCCCCUAUAUUAAUAGAAUAUAAUAUAGACUACCAUUCAUUAUCCAAAUAAAUAUUAUUUCAUACAAGGCUUUAAAAUUU-GAUAAGCAUCAGAUGGCAAAUUUUUU-CUCGGUGUAUUGG .....(((..(((((((((((((....((((..((((((((.(((.((((((...((((((((........)))))))).................(((((.....))))).......)))))).)))..)-)))))))....))))....)))))))-).)))))..))). ( -36.50, z-score = -2.74, R) >droSec1.super_2 2573326 152 + 7591821 UCUUCCACUGCACUUGAAAAAACGUUCUAUAUAAUUUAUCAUUUAAGA----------UAUUGAUGGAAUAUUUAAUUCACUACCACACAUC---CGAA--AAUUUAA----AUACAAGACCUUUAAAUUAAGAUAAGCAUCAAAUAGCACAUUUUUU-CUCAGUGUAUUGG ....(((.((((((.(((((((.((.((((......((((......))----------))((((((((((.....)))).............---....--(((((((----(.........))))))))........)))))))))).)).))))))-)..)))))).))) ( -28.10, z-score = -3.47, R) >droSim1.chr3L 2108793 153 + 22553184 UCUUCCAGUGCACUGGAAAAAACGUUCUAUAAUGUUUAUCAAUUAAGA----------UAUUGCUGGAAUAUUUUAUUCACUAACACACAUC---UAAA--AAUUUCA----AUACAAGACCUUUAAGUUGAGAUAAGCAUCAGAUAUCAAAUUUUUUUCUCAGUGUAUUGG ....((((((((((((((((((.(((..((((((((((((...(((((----------((((.....)))))))))................---....--....(((----((.............))))))))))))))....)))..)))))))))).))))))))))) ( -35.02, z-score = -3.75, R) >consensus UCUUCCAGUGCACUGGAAAAAACGUUAUAUAUGGUUUAUCAAUUAAGA__________UAUUGAUGGAAUAUUUUAUUCACUACCACACAUC___CAAA__AAUUUUA____AUACAAGACCUUUAAAUUGAGAUAAGCAUCAGAUAGCAAAUUUUUU_CUCAGUGUAUUGG ....((((((((((((((((((.((..(((...(((((((..........................((((.....))))......................(((((((...............)))))))..))))))).....)))..)).))))))..)))))))))))) (-15.35 = -17.29 + 1.94)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:58:02 2011