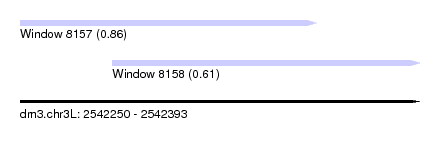
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 2,542,250 – 2,542,393 |
| Length | 143 |
| Max. P | 0.859002 |

| Location | 2,542,250 – 2,542,356 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 71.41 |
| Shannon entropy | 0.54495 |
| G+C content | 0.42120 |
| Mean single sequence MFE | -25.72 |
| Consensus MFE | -12.22 |
| Energy contribution | -13.53 |
| Covariance contribution | 1.31 |
| Combinations/Pair | 1.37 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.859002 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
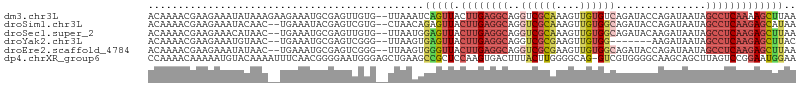
>dm3.chr3L 2542250 106 + 24543557 ACAAAACGAAGAAAUAUAAAGAAGAAAUGCGAGUUGUG--UUAAAUCAGUUACUUGAGGCAGGUCGCAAAGUUGUGUCAGAUACCAGAUAAUAGCCUCAAAAGCUUAA .....................(((...(((((.((((.--((((.........)))).)))).)))))..((((((((........))).)))))........))).. ( -17.90, z-score = -0.19, R) >droSim1.chr3L 2097943 104 + 22553184 ACAAAACGAAGAAAUACAAC--UGAAAUACGAGUCGUG--CUAACAGAGUUACUUGAGGCAGGUCGCAAAGUUGUGGCAGAUACCAGAUAAUAGCCUCAAGAGCAUAA ...................(--((...((((...))))--....))).(((.((((((((..((((((....)))))).(....)........))))))))))).... ( -23.70, z-score = -1.70, R) >droSec1.super_2 2563066 104 + 7591821 ACAAAACGAAGAAACAUAAC--UGAAAUGCGAGUUGUG--UUAAUGGAGUUACUUGAGGCAGGUCGCAAAGUUGUGGCAGAUACAAGAUAAUAGCCUCAAGAGCUUAA ............((((((((--(........)))))))--))....(((((.((((((((..((((((....))))))..((.....))....))))))))))))).. ( -30.80, z-score = -3.64, R) >droYak2.chr3L 14799786 97 - 24197627 ACAAAACGAAGAAAUGUAAC--UGAAAUGCGAGUCGGG--UUAAGUGAGUUACUUGAGGCAGGUCGCGAAGUUGUGG-------AAGAUAAUAGCCUCAAGAGCUUAC ....(((...((..((((..--.....))))..))..)--))..(((((((.((((((((..(((............-------..)))....))))))))))))))) ( -23.94, z-score = -1.61, R) >droEre2.scaffold_4784 2555099 104 + 25762168 ACAAAACGAAGAAAUAUAAC--UGAAAUGCGAGUCGGG--UUAAGUGGGUUACUUGAGGCAGGUCGCGAAGUUGUGGCAGAUACCAGAUAAUAGCCUCAAGAGCUUAA ...................(--(((........)))).--.....((((((.((((((((..((((((....)))))).(....)........)))))))))))))). ( -25.30, z-score = -1.16, R) >dp4.chrXR_group6 12533086 107 + 13314419 CCAAAACAAAAAUGUACAAAAUUUCAACGGGGAAUGGGAGCUGAAGCCGCUCCAAGUGACUUUACUUGGGGCAG-GUCGUGGGGCAAGCAGCUUAGUCCGGAAUGGAA (((..(((....)))...............(((....((((((..(((((((((((((....))))))))))..-.(....))))...))))))..)))....))).. ( -32.70, z-score = -1.24, R) >consensus ACAAAACGAAGAAAUAUAAC__UGAAAUGCGAGUCGGG__UUAAAUGAGUUACUUGAGGCAGGUCGCAAAGUUGUGGCAGAUACCAGAUAAUAGCCUCAAGAGCUUAA ..............................................(((((.((((((((..((((((....))))))...............))))))))))))).. (-12.22 = -13.53 + 1.31)


| Location | 2,542,283 – 2,542,393 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 71.58 |
| Shannon entropy | 0.50507 |
| G+C content | 0.42278 |
| Mean single sequence MFE | -28.67 |
| Consensus MFE | -12.59 |
| Energy contribution | -12.35 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.61 |
| Mean z-score | -1.63 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.610056 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 2542283 110 + 24543557 ------UUGUGUUAAAUCAGUU-ACUUGAGGCAGGUCGCAAAGUUGUGUCAGAUACCAGAUAAUAGCCUCAAAAGCUUAAUUUGCUAA-UAUACAGUAGUAGUAAGUAAGAGAGUGUG ------...............(-(((((((((..(.((((....)))).).(....)........))))).....((((.(((((((.-(((...))).))))))))))).))))).. ( -20.30, z-score = 0.18, R) >droSim1.chr3L 2097974 107 + 22553184 ------UCGUGCUAACAGAGUU-ACUUGAGGCAGGUCGCAAAGUUGUGGCAGAUACCAGAUAAUAGCCUCAAGAGCAUAAUUUGCUAACUAUAUAUUAGUUGUAAG----AGAGUGUG ------.((..((......(((-.((((((((..((((((....)))))).(....)........)))))))))))....(((((.(((((.....))))))))))----..))..)) ( -30.40, z-score = -2.15, R) >droSec1.super_2 2563097 107 + 7591821 ------UUGUGUUAAUGGAGUU-ACUUGAGGCAGGUCGCAAAGUUGUGGCAGAUACAAGAUAAUAGCCUCAAGAGCUUAAUUUGCUAACUAUAUAUUAGUAGUAAG----AGAGUGUG ------...........(((((-.((((((((..((((((....))))))..((.....))....)))))))))))))..(((((((.(((.....))))))))))----........ ( -28.70, z-score = -2.02, R) >droYak2.chr3L 14799817 99 - 24197627 ------UCGGGUUAAGUGAGUU-ACUUGAGGCAGGUCGCGAAGUUGUGG-------AAGAUAAUAGCCUCAAGAGCUUACUUUCCUGA-UAUAUAGUAGUGGUAAG----AGAGAGUG ------(((((..(((((((((-.((((((((..(((............-------..)))....))))))))))))))))).)))))-.................----........ ( -31.44, z-score = -3.36, R) >droEre2.scaffold_4784 2555130 106 + 25762168 ------UCGGGUUAAGUGGGUU-ACUUGAGGCAGGUCGCGAAGUUGUGGCAGAUACCAGAUAAUAGCCUCAAGAGCUUAAUUUGCUAA-UAUAUAGUGGCAGUAAG----AGAGAGUG ------..........((((((-.((((((((..((((((....)))))).(....)........))))))))))))))..((((((.-.......))))))....----........ ( -27.10, z-score = -1.03, R) >dp4.chrXR_group6 12533126 102 + 13314419 CUGAAGCCGCUCCAAGUGACUUUACUUGGGGCAGGUCGUGGGGCAAGC---------AGCU--UAGUCCGGAAUGGAAUACAUAUGCA-CAUGUGUGUGUGGCAAG----AGAAAGUC .....(((((((((((((....)))))))))).((.(...((((....---------.)))--).).))..........((((((((.-...)))))))))))...----........ ( -34.10, z-score = -1.42, R) >consensus ______UCGUGUUAAGUGAGUU_ACUUGAGGCAGGUCGCAAAGUUGUGGCAGAUACCAGAUAAUAGCCUCAAGAGCUUAAUUUGCUAA_UAUAUAGUAGUAGUAAG____AGAGAGUG ...................(((..((((((((..((((((....))))))...............))))))))))).....((((((.........))))))................ (-12.59 = -12.35 + -0.24)

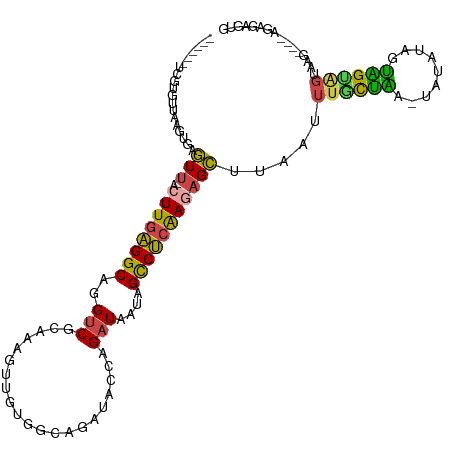
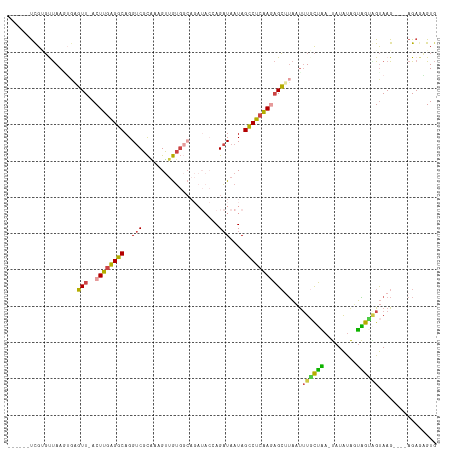
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:58:00 2011