| Sequence ID | dm3.chr3L |
|---|---|
| Location | 2,541,808 – 2,541,904 |
| Length | 96 |
| Max. P | 0.878877 |

| Location | 2,541,808 – 2,541,904 |
|---|---|
| Length | 96 |
| Sequences | 10 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 60.37 |
| Shannon entropy | 0.80049 |
| G+C content | 0.44980 |
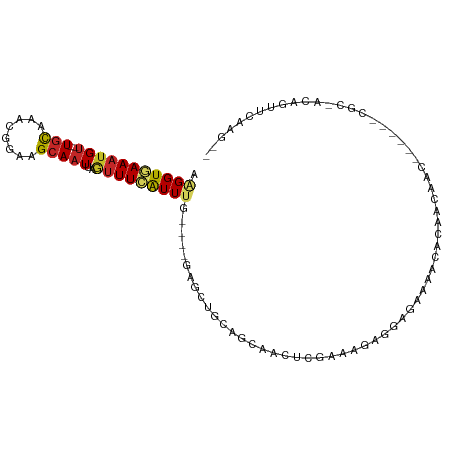
| Mean single sequence MFE | -24.06 |
| Consensus MFE | -6.93 |
| Energy contribution | -7.05 |
| Covariance contribution | 0.12 |
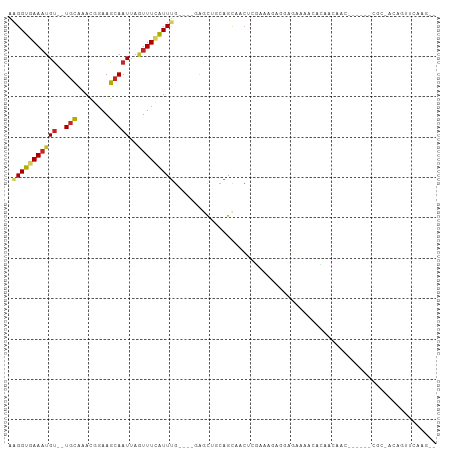
| Combinations/Pair | 1.42 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.29 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.878877 |
| Prediction | RNA |
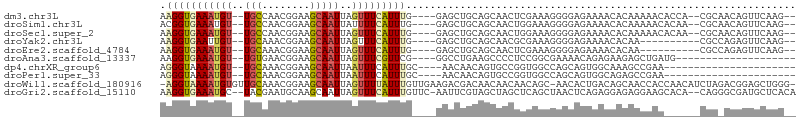
Download alignment: ClustalW | MAF
>dm3.chr3L 2541808 96 + 24543557 AAGGUGAAAUGU--UGCCAACGGAAGCAAUUAGUUUCAUUUG----GAGCUGCAGCAACUCGAAAGGGGAGAAAACACAAAAACACCA--CGCAACAGUUCAAG-- ....((((.(((--(((....((..((...(((((((....)----))))))..))..(((....))).................)).--.)))))).))))..-- ( -24.70, z-score = -2.11, R) >droSim1.chr3L 2097501 96 + 22553184 ACGGUGAAAUGU--UGCCAACGGAAGCAAUUAUUUUCAUUUG----GAGCUGCAGCAACUGGAAAGGGGAGAAAACACAAAAACACAA--CGCAACAGUUCAAG-- ..(((((((.((--(((........)))))...)))))))..----((((((..((..((....))(........)............--.))..))))))...-- ( -19.40, z-score = -0.59, R) >droSec1.super_2 2562626 96 + 7591821 AAGGUGAAAUGU--UGCCAACGGAAGCAAUUAGUUUCAUUUG----GAGCUGCAGCAACUGGAAAGGGGAGAAAACACAAAAACACAA--CGCAACAGUUCAAG-- .(((((((((((--(((........)))))..))))))))).----((((((..((..((....))(........)............--.))..))))))...-- ( -20.90, z-score = -0.98, R) >droYak2.chr3L 14799338 88 - 24197627 AAGGUGAAUUGU--UGCAAACGGAAGCAAUUAGUUUCAUUUG----GAGCUGCAGCAACGCGAAAGGGGAGAAAACACAA----------CGCCAGAGUUCAAG-- ..((((..((((--(((...(....).....((((((....)----))))))))))))(.(....).)............----------))))..........-- ( -23.30, z-score = -1.88, R) >droEre2.scaffold_4784 2554676 88 + 25762168 AAGGUGAAAUGU--UGCAAACGGAAGCAAUUAGUUUCAUUUG----GAGCUGCAGCAACUCGAAAGGGGAGAAAACACAA----------CGCCAGAGUUCAAG-- ..((((...(((--(((...(....).....((((((....)----))))))))))).(((....)))............----------))))..........-- ( -24.60, z-score = -2.50, R) >droAna3.scaffold_13337 557132 80 + 23293914 AAGGUGAAAUGU--UGUGAACGGAAGCAAUUAGUUUCGUUCG----GGCCUGAAGCCCCUCCGGCGAAAACAGAGAAGAGCUGAUG-------------------- ..(((......(--((((((((.((((.....)))))))))(----(((.....))))...........))))......)))....-------------------- ( -18.60, z-score = 0.11, R) >dp4.chrXR_group6 12532617 78 + 13314419 AGGGUAAAAUGU--UGCAAACGGAAGCAAUUAAUUUCAUUUGC----AACAACAGUGCCGGUGGCCAGCAGUGGCAAAGCCGAA---------------------- ..((((...(((--((((((.((((........)))).)))))----))))....))))(((.((((....))))...)))...---------------------- ( -28.20, z-score = -3.06, R) >droPer1.super_33 552304 78 + 967471 AGGGUAAAAUGU--UGCAAACGGAAGCAAUUAAUUUCAUUUGC----AACAACAGUGCCGGUGGCCAGCAGUGGCAGAGCCGAA---------------------- ..((((...(((--((((((.((((........)))).)))))----))))....))))(((.((((....))))...)))...---------------------- ( -28.20, z-score = -3.07, R) >droWil1.scaffold_180916 2141926 103 + 2700594 -AGGUAAAAUGUGUUGCAAACGGAAGCAAUUAGUUUUAUUUGUUGAAGACGACAACAACAACAGC-AACACUGACAGCAACCACCAACAUCUAGACGGAGCUGGG- -.(((.....(((((((.....(((((.....)))))..((((((....))))))........))-)))))........))).(((...(((....)))..))).- ( -22.52, z-score = -0.95, R) >droGri2.scaffold_15110 24370783 101 - 24565398 AAGGUGAAAUGC--UACGAAUGCAAGCAAUUAGUUUCAUUUGUUC-AAUUCGUAGCUAGCUCAGCUAACUCAGAGGAGAGGAAGCACA--CAGGGCGAUGCUCACA ...((((...((--(((((((...(((((..........))))).-.)))))))))..((((.(((..(((......)))..)))...--..)))).....)))). ( -30.20, z-score = -2.01, R) >consensus AAGGUGAAAUGU__UGCAAACGGAAGCAAUUAGUUUCAUUUG____GAGCUGCAGCAACUCGAAAGAGGAGAAAACACAACAAC______CGC_ACAGUUCAAG__ .(((((((((....(((........)))....)))))))))................................................................. ( -6.93 = -7.05 + 0.12)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:57:58 2011