| Sequence ID | dm3.chr3L |
|---|---|
| Location | 2,521,232 – 2,521,333 |
| Length | 101 |
| Max. P | 0.836590 |

| Location | 2,521,232 – 2,521,333 |
|---|---|
| Length | 101 |
| Sequences | 7 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 68.08 |
| Shannon entropy | 0.60437 |
| G+C content | 0.50284 |
| Mean single sequence MFE | -15.94 |
| Consensus MFE | -7.48 |
| Energy contribution | -7.06 |
| Covariance contribution | -0.43 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.836590 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
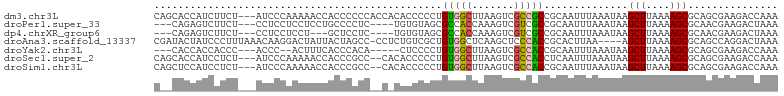
>dm3.chr3L 2521232 101 + 24543557 CAGCACCAUCUUCU---AUCCCAAAAACCACCCCCCACCACACCCCCUGUGGCUUAAGUCGCCGCCGCAAUUUAAAUAAGCUUAAAAGCGCAGCGAAGACCAAA ........(((((.---....................(((((.....)))))........((.((.((..(((((......))))).)))).)))))))..... ( -16.50, z-score = -2.21, R) >droPer1.super_33 524188 94 + 967471 ---CAGAGUCUUCU---CCUCCUCCUCCUGCCCCUC----UGUGUAGCGCCACCAAAGUCGUCGCCGCAAUUUAAAUAAGCUUAAAAGCGCAACGAAGACUAAA ---...(((((((.---...................----..((..((((.((....)).).)))..))..........(((....))).....)))))))... ( -18.10, z-score = -1.81, R) >dp4.chrXR_group6 12505121 91 + 13314419 ---CAGAGUCUUCU---CCUCCUCCU---GCUCCUC----UGUGUAGCGCCACCAAAGUCGUCGCCGCAAUUUAAAUAAGCUUAAAAGCGCAACGAAGACUAAA ---...(((((((.---.........---.......----..((..((((.((....)).).)))..))..........(((....))).....)))))))... ( -18.10, z-score = -1.36, R) >droAna3.scaffold_13337 541483 99 + 23293914 CGAUACUAUCCCUUUAAACAAGGACUAUUACUAGCC-CCUCUGUCGCUGUGGCUCAAGCUCCCACCGCACUUAA----AGCUUAAAAGCGCAGCCAGGACUAAA ..........((((.....))))........(((((-...((((.((.((((.........)))).))......----.(((....)))))))...)).))).. ( -17.90, z-score = 0.51, R) >droYak2.chr3L 14778056 91 - 24197627 ---CACCACCACCC---ACCC--ACUUUCACCCACA-----CUCCCCUGUGGCUUAAGUCGCCACCGCAAUUUAAAUAAGCUUAAAAGCGCAGCGAAGACCAAA ---...........---....--.(((....(((((-----......)))))...)))((((...(((..(((((......))))).)))..))))........ ( -14.00, z-score = -1.93, R) >droSec1.super_2 2542217 99 + 7591821 CAGCACCAUCCUCU---AUCCCAAAAACCACCCGCC--CACACCCCCUGUGGCUUAAGUCGCCACCUCAAUUUAAAUAAGCUUAAAAGCGCAGCGAAGACCAAA ..............---...............(((.--..........(((((.......)))))..............(((....)))...)))......... ( -13.00, z-score = -1.25, R) >droSim1.chr3L 2075139 99 + 22553184 CAGCUCCAUCCUCU---AUCCCAAAAACCACCCGCC--CACACCCCCUGUGGCUUAAGUCGCCACCGCAAUUUAAAUAAGCUUAAAAGCGCAGCGAAGACCAAA ...........(((---................(((--.(((.....)))))).....((((...(((..(((((......))))).)))..)))))))..... ( -14.00, z-score = -1.12, R) >consensus C__CACCAUCCUCU___ACCCCAACUACCACCCCCC__C_CACCCCCUGUGGCUUAAGUCGCCACCGCAAUUUAAAUAAGCUUAAAAGCGCAGCGAAGACCAAA ................................................(((((.......)))))..............(((....)))............... ( -7.48 = -7.06 + -0.43)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:57:55 2011