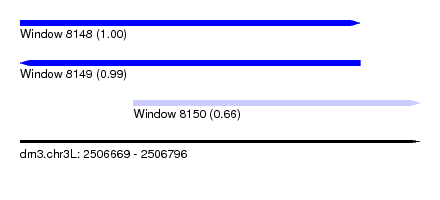
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 2,506,669 – 2,506,796 |
| Length | 127 |
| Max. P | 0.996548 |

| Location | 2,506,669 – 2,506,777 |
|---|---|
| Length | 108 |
| Sequences | 7 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 78.37 |
| Shannon entropy | 0.41726 |
| G+C content | 0.58558 |
| Mean single sequence MFE | -47.06 |
| Consensus MFE | -35.29 |
| Energy contribution | -36.39 |
| Covariance contribution | 1.10 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.75 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.996548 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 2506669 108 + 24543557 ----GAGACACAUGCGGUGAUUAUGCGCUGCCGGCCGUCAGGUCCACAGGAUCCUCCAUUCCGGUGGGAUGGAGUCUCCUUCUGGCCGCGGCCGGG--GUCUUAAA-GUCAAAUA ----(((((....((((((......))))))(((((((..((.(((.((((..(((((((((...)))))))))..))))..))))))))))))..--)))))...-........ ( -51.90, z-score = -3.59, R) >droSim1.chr3L 2060523 108 + 22553184 ----GAGACACAUGCGGUGAUUAUGCGCUGCCGGCCGUCAGGUCCACAGGAUCCUCCAUUCCAGUGGGAUGGAGUCUCCUCCUGGCCGCGGCCGGG--GUCUUAAA-GUCAAAUA ----(((((....((((((......))))))(((((((..((((...((((..(((((((((...)))))))))..))))...)))))))))))..--)))))...-........ ( -51.60, z-score = -3.65, R) >droSec1.super_2 2527947 108 + 7591821 ----GAGACACAUGCGGUGAUUAUGCGCUGCCGGCCGUCAGGUCCACAGGAUCCUCCAUUCCGGUGGGAUGGAGUCUCCUCCUGGCCGCGGCCGGG--GUCUUAAA-GUCAAAUA ----(((((....((((((......))))))(((((((..((((...((((..(((((((((...)))))))))..))))...)))))))))))..--)))))...-........ ( -51.60, z-score = -3.29, R) >droYak2.chr3L 14763155 108 - 24197627 ----GAGACACAUGCGGUGAUUAUGCGCUGCCGGCCGUCGGAUCCACGGACUCCUCCAUUCCGGUGGAAUGGAUUAUCCUUGUGGCCACGGCCGGG--GUCUUAAA-GUCAAAUA ----(((((....((((((......))))))(((((((.((..(((((((....((((((((...))))))))...)))..)))))))))))))..--)))))...-........ ( -46.40, z-score = -2.82, R) >droEre2.scaffold_4784 2519378 108 + 25762168 ----GAGACACAUGCGGUGAUUAUGCGCUGCCGGCCGCCGGGUCCACGGGAUCCUCCAUUCGGUUGGGAUGGAGUCUCCUUGUGGCCGCGGCCGGG--GUCUUAAA-GUCAAAUA ----(((((....((((((......))))))(((((((..((.((((((((..((((((((.....))))))))..))).))))))))))))))..--)))))...-........ ( -54.90, z-score = -3.63, R) >droAna3.scaffold_13337 529026 95 + 23293914 ----AAUGCACAUGCGGUGAAUAUGCGCUGCCGGCCGUCGACUCCCGGGGCGCCUC-------------UUGGGCACCCACCUGGCCACGGCCGGG--GUCUUAAA-GUCAAAUA ----...((....((((((......))))))..))....((((....(((.(((..-------------...))).))).((((((....))))))--.......)-)))..... ( -37.20, z-score = -0.02, R) >droWil1.scaffold_180916 2092989 111 + 2700594 GACUAAUGCACAUGCGGUGAAUAUGCGCUGCCGGACAUUGGGCUCGAUUUGUGGUC-ACAUGGUUAG--UGUGUCCACUUCUUGGCCACGACCGGGUUGUCUUAAAAGUCAAAC- ((((.........((((((......))))))(((...((((((.(((...((((.(-((((.....)--)))).))))...)))))).))))))............))))....- ( -35.80, z-score = -1.06, R) >consensus ____GAGACACAUGCGGUGAUUAUGCGCUGCCGGCCGUCGGGUCCACAGGAUCCUCCAUUCCGGUGGGAUGGAGUCUCCUCCUGGCCGCGGCCGGG__GUCUUAAA_GUCAAAUA ....(((((....((((((......))))))(((((((..((((...(((...((((((((.....))))))))...)))...)))))))))))....)))))............ (-35.29 = -36.39 + 1.10)
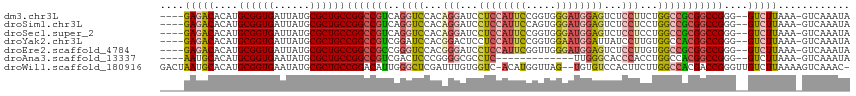
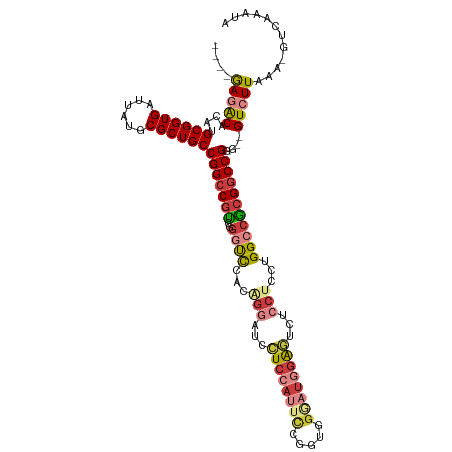
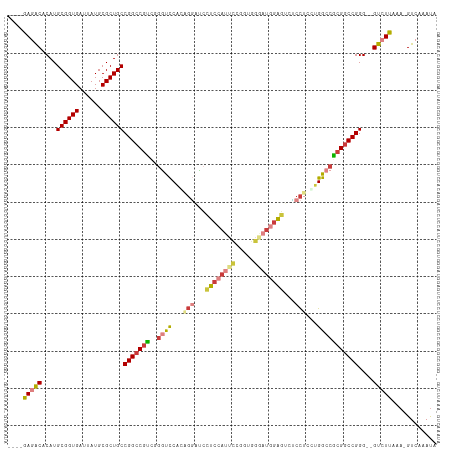
| Location | 2,506,669 – 2,506,777 |
|---|---|
| Length | 108 |
| Sequences | 7 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 78.37 |
| Shannon entropy | 0.41726 |
| G+C content | 0.58558 |
| Mean single sequence MFE | -41.70 |
| Consensus MFE | -25.82 |
| Energy contribution | -27.01 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.987166 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 2506669 108 - 24543557 UAUUUGAC-UUUAAGAC--CCCGGCCGCGGCCAGAAGGAGACUCCAUCCCACCGGAAUGGAGGAUCCUGUGGACCUGACGGCCGGCAGCGCAUAAUCACCGCAUGUGUCUC---- ........-....((((--.(((((((((((((..((((..((((((((....)).))))))..)))).))).)).).)))))))..(((.........)))....)))).---- ( -44.90, z-score = -3.23, R) >droSim1.chr3L 2060523 108 - 22553184 UAUUUGAC-UUUAAGAC--CCCGGCCGCGGCCAGGAGGAGACUCCAUCCCACUGGAAUGGAGGAUCCUGUGGACCUGACGGCCGGCAGCGCAUAAUCACCGCAUGUGUCUC---- ........-....((((--.(((((((((((((.(.(((..((((((((....)).))))))..)))).))).)).).)))))))..(((.........)))....)))).---- ( -43.70, z-score = -2.47, R) >droSec1.super_2 2527947 108 - 7591821 UAUUUGAC-UUUAAGAC--CCCGGCCGCGGCCAGGAGGAGACUCCAUCCCACCGGAAUGGAGGAUCCUGUGGACCUGACGGCCGGCAGCGCAUAAUCACCGCAUGUGUCUC---- ........-....((((--.(((((((((((((.(.(((..((((((((....)).))))))..)))).))).)).).)))))))..(((.........)))....)))).---- ( -43.80, z-score = -2.43, R) >droYak2.chr3L 14763155 108 + 24197627 UAUUUGAC-UUUAAGAC--CCCGGCCGUGGCCACAAGGAUAAUCCAUUCCACCGGAAUGGAGGAGUCCGUGGAUCCGACGGCCGGCAGCGCAUAAUCACCGCAUGUGUCUC---- ........-....((((--.((((((((((((((..((((..((((((((...))))))))...))))))))..)).))))))))..(((.........)))....)))).---- ( -43.70, z-score = -3.12, R) >droEre2.scaffold_4784 2519378 108 - 25762168 UAUUUGAC-UUUAAGAC--CCCGGCCGCGGCCACAAGGAGACUCCAUCCCAACCGAAUGGAGGAUCCCGUGGACCCGGCGGCCGGCAGCGCAUAAUCACCGCAUGUGUCUC---- ........-....((((--.((((((((((((((..(((..((((((.(.....).))))))..))).)))).))..))))))))..(((.........)))....)))).---- ( -44.40, z-score = -3.06, R) >droAna3.scaffold_13337 529026 95 - 23293914 UAUUUGAC-UUUAAGAC--CCCGGCCGUGGCCAGGUGGGUGCCCAA-------------GAGGCGCCCCGGGAGUCGACGGCCGGCAGCGCAUAUUCACCGCAUGUGCAUU---- ........-........--.((((((((((((..(.(((((((...-------------..))))))))..).))).))))))))..((((((.........))))))...---- ( -41.20, z-score = -1.59, R) >droWil1.scaffold_180916 2092989 111 - 2700594 -GUUUGACUUUUAAGACAACCCGGUCGUGGCCAAGAAGUGGACACA--CUAACCAUGU-GACCACAAAUCGAGCCCAAUGUCCGGCAGCGCAUAUUCACCGCAUGUGCAUUAGUC -.............(((...((((.((((((...((.((((.((((--.......)))-).))))...))..)))..))).))))..((((((.........))))))....))) ( -30.20, z-score = -1.10, R) >consensus UAUUUGAC_UUUAAGAC__CCCGGCCGCGGCCAGGAGGAGACUCCAUCCCACCGGAAUGGAGGAUCCCGUGGACCCGACGGCCGGCAGCGCAUAAUCACCGCAUGUGUCUC____ ....................(((((((((((((...((...((((((.(.....).))))))...))..)))..))).)))))))..((((((.........))))))....... (-25.82 = -27.01 + 1.20)

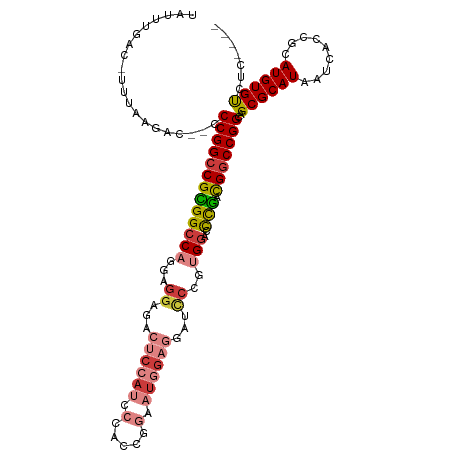
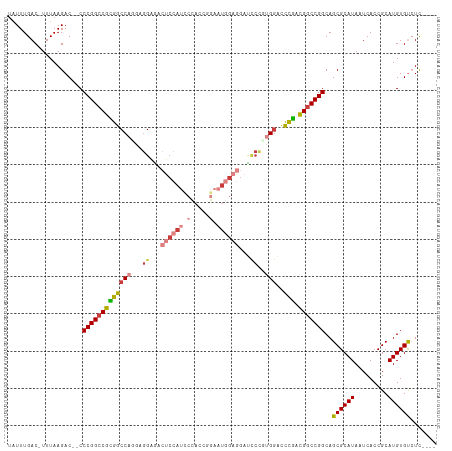
| Location | 2,506,705 – 2,506,796 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 79.35 |
| Shannon entropy | 0.39040 |
| G+C content | 0.54008 |
| Mean single sequence MFE | -32.63 |
| Consensus MFE | -20.90 |
| Energy contribution | -22.07 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.42 |
| Mean z-score | -1.45 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.656166 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 2506705 91 + 24543557 GGUCCACAGGAUCCUCCAUUCCGGUGGGAUGGAGUCUCCUUCUGGCCGCGGCCGGG--GUCUUAAA-GUCAAAUAGAGACAGAGUGAUAGAGAG ....(((((((..(((((((((...)))))))))..))))((((((....))))))--(((((...-........)))))...)))........ ( -34.20, z-score = -1.28, R) >droSim1.chr3L 2060559 91 + 22553184 GGUCCACAGGAUCCUCCAUUCCAGUGGGAUGGAGUCUCCUCCUGGCCGCGGCCGGG--GUCUUAAA-GUCAAAUAGAGACAGAGUGAUAGAGAG ....(((((((..(((((((((...)))))))))..))))((((((....))))))--(((((...-........)))))...)))........ ( -36.90, z-score = -2.21, R) >droSec1.super_2 2527983 91 + 7591821 GGUCCACAGGAUCCUCCAUUCCGGUGGGAUGGAGUCUCCUCCUGGCCGCGGCCGGG--GUCUUAAA-GUCAAAUAGAGACAGAGUGAUAGAGAG ....(((((((..(((((((((...)))))))))..))))((((((....))))))--(((((...-........)))))...)))........ ( -36.90, z-score = -1.85, R) >droYak2.chr3L 14763191 91 - 24197627 GAUCCACGGACUCCUCCAUUCCGGUGGAAUGGAUUAUCCUUGUGGCCACGGCCGGG--GUCUUAAA-GUCAAAUAGAGACAGAGUGAUAGAGAG ....((((((((((((((((((...))))))))..........(((....))))))--))))....-(((.......)))...)))........ ( -31.20, z-score = -1.57, R) >droEre2.scaffold_4784 2519414 91 + 25762168 GGUCCACGGGAUCCUCCAUUCGGUUGGGAUGGAGUCUCCUUGUGGCCGCGGCCGGG--GUCUUAAA-GUCAAAUAGAGACAGAGUGAUAGAGAG ((.((((((((..((((((((.....))))))))..))).)))))))(((((....--))).....-(((.......)))...))......... ( -32.00, z-score = -0.83, R) >droWil1.scaffold_180916 2093029 87 + 2700594 GGCUCGAUUUGUGGUC-ACAUGGUUAG--UGUGUCCACUUCUUGGCCACGACCGGGUUGUCUUAAAAGUCAAACUAGAGUAGACAGAAAG---- (((.(((...((((.(-((((.....)--)))).))))...)))))).........((((((....((.....)).....))))))....---- ( -24.60, z-score = -0.99, R) >consensus GGUCCACAGGAUCCUCCAUUCCGGUGGGAUGGAGUCUCCUCCUGGCCGCGGCCGGG__GUCUUAAA_GUCAAAUAGAGACAGAGUGAUAGAGAG ....(((((((..((((((((.....))))))))..))))((((((....))))))..(((((............)))))...)))........ (-20.90 = -22.07 + 1.17)

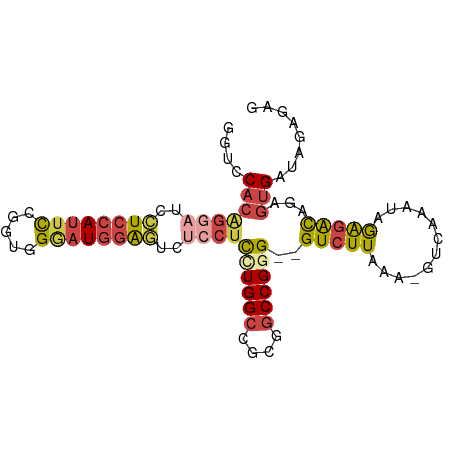
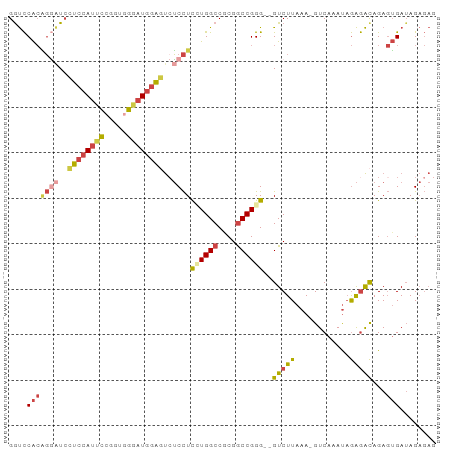
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:57:53 2011