| Sequence ID | dm3.chr3L |
|---|---|
| Location | 2,408,252 – 2,408,347 |
| Length | 95 |
| Max. P | 0.975607 |

| Location | 2,408,252 – 2,408,347 |
|---|---|
| Length | 95 |
| Sequences | 11 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 80.19 |
| Shannon entropy | 0.39898 |
| G+C content | 0.46399 |
| Mean single sequence MFE | -29.56 |
| Consensus MFE | -20.50 |
| Energy contribution | -19.96 |
| Covariance contribution | -0.53 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.69 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.93 |
| SVM RNA-class probability | 0.975607 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
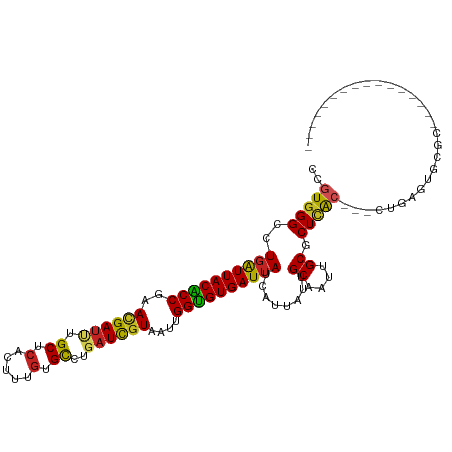
>dm3.chr3L 2408252 95 + 24543557 CUGUGGGCCUGAUUACACCGAACGAUUUGCUCACUUUGUGCCUGAUCGUAAUUGGUGUGAUUAUCAUUAUUGCAAUUGCUCUCAC---CUGAGUGCGC---------------- .((..(...((((((((((((((((((.((.(.....).))..))))))..))))))))))))......)..))...((.(((..---..))).))..---------------- ( -29.90, z-score = -3.11, R) >droGri2.scaffold_15110 24248791 90 - 24565398 CCGUGGGUCUGAUUACGCCCAAUGAUUUGCUCACGUUGUGUCUGAUUGUAAUUGGUGUGAUUAUCAUUGUUGCAAUUGCGCUCAG---CUGAA--------------------- ..((((((..(((((.......))))).))))))((((.((.(((((((((((((((....)))))..)))))))))).)).)))---)....--------------------- ( -24.90, z-score = -1.26, R) >droMoj3.scaffold_6680 2237912 93 + 24764193 CCGUGGGCCUGAUUACACCGAACGAUUUGCUCACAUUGUGUCUGAUCGUAAUUGGUGUGAUUAUCAUUGUGGCAAUUGCGCUCAC---CUGAGCGC------------------ (((..(...((((((((((((((((((....(((...)))...))))))..))))))))))))...)..))).....((((((..---..))))))------------------ ( -35.60, z-score = -3.56, R) >droVir3.scaffold_10322 183962 101 + 456481 CCGUGGGCCUGAUUACGCCCAAUGAUUUGCUCACAUUGUGCCUGAUCGUAAUUGGUGUGAUUAUCAUUGUGGCAAUAGCGCUCGC---CUGAACACACACACAC---------- ..(((((((((.....((((((((((....(((((((.(((......)))...)))))))..))))))).)))..))).))))))---................---------- ( -29.10, z-score = -0.82, R) >droWil1.scaffold_180916 1967004 89 + 2700594 CAGUGGGCCUGAUUACACCUAAUGAUUUGCUCACUUUAUGUUUGAUCGUAAUUGGUGUGAUUAUCAUUAUAGCAAUUGCUCUCAA---CUGA---------------------- .(((((((..(((((.......))))).)))))))......((((..(((((((.((((((....)))))).)))))))..))))---....---------------------- ( -23.70, z-score = -2.48, R) >droPer1.super_33 390403 114 + 967471 CCGUGGGCCUGAUUACACCCAACGAUUUGCUCACUUUGUGCCUCAUUGUAAUUGGUGUGAUUAUCAUUAUUGCAAUUGCUCUUGCUGCCUGAAAGAUGUGAUCAUCACAUACAC ..((((((.((((((((((..(((((..((.(.....).))...)))))....))))))))))........((((......)))).)))......(((((.....))))).))) ( -27.90, z-score = -1.09, R) >dp4.chrXR_group6 12369265 114 + 13314419 CCGUGGGCCUGAUUACACCCAACGAUUUGCUCACUUUGUGCCUCAUUGUAAUUGGUGUGAUUAUCAUUAUUGCAAUUGCUCUCGCUGCCUGAAAGAUGUGAUCAUCACAUACAC ..((((((.((((((((((..(((((..((.(.....).))...)))))....))))))))))........((....)).......)))......(((((.....))))).))) ( -26.20, z-score = -0.51, R) >droAna3.scaffold_13337 452594 99 + 23293914 CCGUGGGCCUGGUUACGCCGAACGACCUGCUCACUCUGUGCCUGAUCAUAAUUGGCGUGAUUAUAAUUAUUGCAAUUGCGCUGGC---CUGAUGGGGAGAGU------------ ..((((((..((((.........)))).)))))).((.(.(((.((((...(..(((..(((.((.....)).)))..)))..).---.)))).)))).)).------------ ( -30.90, z-score = -0.75, R) >droEre2.scaffold_4784 2423883 95 + 25762168 CUGUGGGCCUGAUUACACCGAACGAUUUGCUCACUUUGUGCCUGAUCGUAAUUGGUGUGAUUAUCAUUAUUGCAAUUGCGCUCGC---CUGAGUGCGC---------------- .((..(...((((((((((((((((((.((.(.....).))..))))))..))))))))))))......)..))...((((((..---..))))))..---------------- ( -32.80, z-score = -3.26, R) >droYak2.chr3L 14669824 95 - 24197627 CCGUGGGCCUGAUUACGCCGAACGAUUUGCUCACUUUGUGCCUGAUCGUAAUUGGUGUGAUUAUCAUUAUUGCAAUUGCGCUCAC---CUGAGUGCGC---------------- ..(..(...((((((((((((((((((.((.(.....).))..))))))..))))))))))))......)..)....((((((..---..))))))..---------------- ( -32.60, z-score = -2.98, R) >droSec1.super_2 2433822 95 + 7591821 CUGUGGGCCUUAUUACACCGAACGAUUUGCUCACUUUGUGCCUGAUCGUAAUUGGUGUGAUUAUCAUUAUUGCAAUUGCGCUCAC---CUGAGUGCGC---------------- .((..(.....((((((((((((((((.((.(.....).))..))))))..))))))))))........)..))...((((((..---..))))))..---------------- ( -31.52, z-score = -3.42, R) >consensus CCGUGGGCCUGAUUACACCGAACGAUUUGCUCACUUUGUGCCUGAUCGUAAUUGGUGUGAUUAUCAUUAUUGCAAUUGCGCUCAC___CUGAGUGCGC________________ ..(((((..((((((((((..((((((.((.(.....).))..))))))....))))))))))........((....)).)))))............................. (-20.50 = -19.96 + -0.53)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:57:37 2011