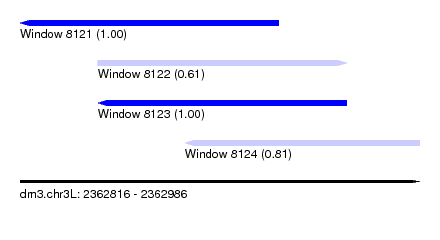
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 2,362,816 – 2,362,986 |
| Length | 170 |
| Max. P | 0.996760 |

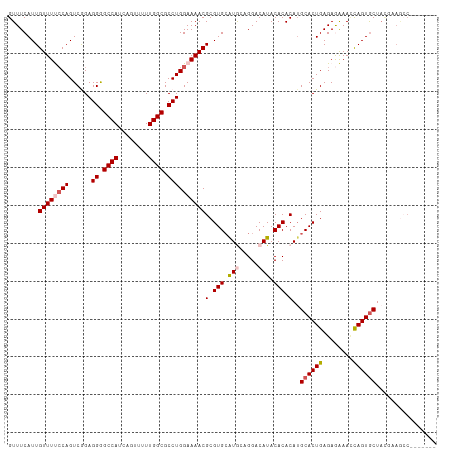
| Location | 2,362,816 – 2,362,926 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 92.45 |
| Shannon entropy | 0.12902 |
| G+C content | 0.50452 |
| Mean single sequence MFE | -34.32 |
| Consensus MFE | -31.44 |
| Energy contribution | -31.92 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.81 |
| SVM RNA-class probability | 0.995529 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 2362816 110 - 24543557 CUGUGUGUAUGUCCUGCAUGCACGCGUUUUCCAGGCGCCAAAAACUGAUGGCCCCUCCGACUGGAAAACAAUGAAACUCAUUUGCAUUUUCCCCCGCUGGUCGGAAAAUG ..(((((((((.....)))))))))((((((((((.((((........)))).)))..((((((((((.((((.....))))....))))))......))))))))))). ( -37.30, z-score = -3.73, R) >droSec1.super_2 2387599 110 - 7591821 AUGUGUGUAUGUCCGCCAUGCACGCGUUUUCCAGGCGCCAAAAACUGAUGGCCCCUCCGACUGGAAAACAAUGAAACUCAUUUGCAUUUUCCCCCGCUCGUCGGAAAAUG ..(((((((((.....)))))))))((((((((((.((((........)))).))......))))))))((((.....))))..((((((((..(....)..)))))))) ( -36.50, z-score = -3.78, R) >droYak2.chr3L 14623176 108 + 24197627 AUGUGUGUAUGUCCUGUAUGCACGCGUUUUCCAGGCGCCAAAA-CUGAUGGCUCCUCUGACUGCGAAACAAUGAAACUCAUUUGCA-UUUGCCCCGCUCGUCGCAAAAUG ..(((((((((.....)))))))))(((((.((((.((((...-....)))).)))..(((.(((....((((.....)))).((.-...))..)))..)))).))))). ( -27.80, z-score = -1.31, R) >droEre2.scaffold_4784 2377755 109 - 25762168 AUGUGUGUAUGUCCUGUACUCACGCGUUUUCCAGGCGCCAAAACCUGAUGGCUCCUCUGACUGGGAAACAAUGAAACUCAUUUGCA-UUUGCCCCGCUCGCCGGAAAAUG ..(((.((((.....)))).))).((((((((.(((((((........))))......((.((((....((((.....)))).((.-...)))))).))))))))))))) ( -33.50, z-score = -2.81, R) >droSim1.chr3L 1925054 110 - 22553184 AUGUGUGUAUGUCCGCCAUGCACGCGUUUUCCAGGCGCCAAAAACUGAUGGCCCCUCCGACUGGAAAACAAUGAAACUCAUUUGCAUUUUCCCCCGCUCGUCGGAAAAUG ..(((((((((.....)))))))))((((((((((.((((........)))).))......))))))))((((.....))))..((((((((..(....)..)))))))) ( -36.50, z-score = -3.78, R) >consensus AUGUGUGUAUGUCCUGCAUGCACGCGUUUUCCAGGCGCCAAAAACUGAUGGCCCCUCCGACUGGAAAACAAUGAAACUCAUUUGCAUUUUCCCCCGCUCGUCGGAAAAUG ..(((((((((.....)))))))))((((((((((.((((........)))).))......))))))))((((.....))))..((((((((..(....)..)))))))) (-31.44 = -31.92 + 0.48)

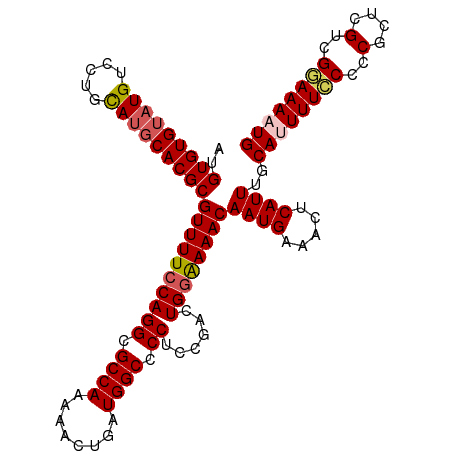
| Location | 2,362,849 – 2,362,955 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 83.68 |
| Shannon entropy | 0.28594 |
| G+C content | 0.50132 |
| Mean single sequence MFE | -32.64 |
| Consensus MFE | -26.26 |
| Energy contribution | -27.14 |
| Covariance contribution | 0.88 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.30 |
| Structure conservation index | 0.80 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.612735 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 2362849 106 + 24543557 GUUUCAUUGUUUUCCAGUCGGAGGGGCCAUCAGUUUUUGGCGCCUGGAAAACGCGUGCAUGCAGGACAUACACACAGGCACUGAGAGAAAACAGUGCUAUGCAGCC------- ........((((((((......((.((((........)))).))))))))))(((((.(((.....))).)))...(((((((........)))))))..))....------- ( -35.20, z-score = -1.55, R) >droSec1.super_2 2387632 98 + 7591821 GUUUCAUUGUUUUCCAGUCGGAGGGGCCAUCAGUUUUUGGCGCCUGGAAAACGCGUGCAUGGCGGACAUACACACAUGCACUGAGAGAAACCAGUGCU--------------- (((((...((((((((......((.((((........)))).))))))))))..((((((((.(......).).))))))).....))))).......--------------- ( -34.70, z-score = -2.01, R) >droYak2.chr3L 14623208 105 - 24197627 GUUUCAUUGUUUCGCAGUCAGAGGAGCCAUCAG-UUUUGGCGCCUGGAAAACGCGUGCAUACAGGACAUACACACAUGCACUGAGAAGAACCAGUGCUACUAAAAC------- ((((...(((..(((..((..(((.((((....-...)))).))).))....))).)))....))))..........((((((........)))))).........------- ( -27.40, z-score = -0.83, R) >droEre2.scaffold_4784 2377787 113 + 25762168 GUUUCAUUGUUUCCCAGUCAGAGGAGCCAUCAGGUUUUGGCGCCUGGAAAACGCGUGAGUACAGGACAUACACACAUGCACUGAGAAGAACCAGUACUACGAAGCCAUUUGCC (((((...((((.((((.(...((((((....))))))...).)))).))))(((((.((...........)).)))))((((........)))).....)))))........ ( -27.00, z-score = -0.02, R) >droSim1.chr3L 1925087 113 + 22553184 GUUUCAUUGUUUUCCAGUCGGAGGGGCCAUCAGUUUUUGGCGCCUGGAAAACGCGUGCAUGGCGGACAUACACACAUGCACUGAGAGAAAUUAGUGCUAUCCAGCCAUAUGCU ........((((((((......((.((((........)))).))))))))))((((..((((((((...........(((((((......)))))))..))).))))))))). ( -38.92, z-score = -2.09, R) >consensus GUUUCAUUGUUUUCCAGUCGGAGGGGCCAUCAGUUUUUGGCGCCUGGAAAACGCGUGCAUGCAGGACAUACACACAUGCACUGAGAGAAACCAGUGCUACGAAGCC_______ ........((((((((......((.((((........)))).))))))))))(.(((.(((.....))).))).)..((((((........))))))................ (-26.26 = -27.14 + 0.88)

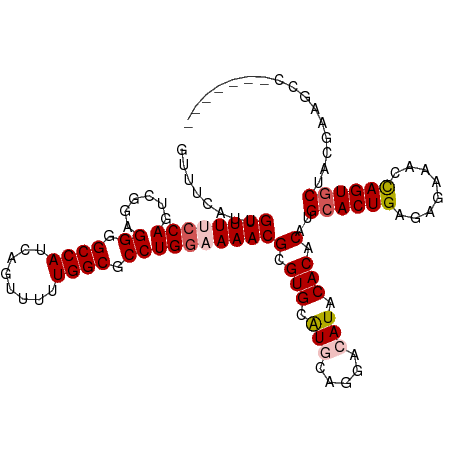
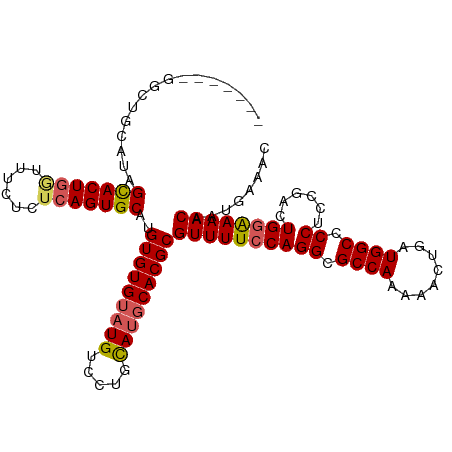
| Location | 2,362,849 – 2,362,955 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 83.68 |
| Shannon entropy | 0.28594 |
| G+C content | 0.50132 |
| Mean single sequence MFE | -35.72 |
| Consensus MFE | -34.08 |
| Energy contribution | -34.32 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.98 |
| SVM RNA-class probability | 0.996760 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 2362849 106 - 24543557 -------GGCUGCAUAGCACUGUUUUCUCUCAGUGCCUGUGUGUAUGUCCUGCAUGCACGCGUUUUCCAGGCGCCAAAAACUGAUGGCCCCUCCGACUGGAAAACAAUGAAAC -------.........((((((........))))))..(((((((((.....)))))))))((((((((((.((((........)))).))......))))))))........ ( -38.10, z-score = -3.53, R) >droSec1.super_2 2387632 98 - 7591821 ---------------AGCACUGGUUUCUCUCAGUGCAUGUGUGUAUGUCCGCCAUGCACGCGUUUUCCAGGCGCCAAAAACUGAUGGCCCCUCCGACUGGAAAACAAUGAAAC ---------------.(((((((......)))))))..(((((((((.....)))))))))((((((((((.((((........)))).))......))))))))........ ( -38.30, z-score = -4.32, R) >droYak2.chr3L 14623208 105 + 24197627 -------GUUUUAGUAGCACUGGUUCUUCUCAGUGCAUGUGUGUAUGUCCUGUAUGCACGCGUUUUCCAGGCGCCAAAA-CUGAUGGCUCCUCUGACUGCGAAACAAUGAAAC -------(((((.((((((((((......)))))))..(((((((((.....)))))))))(((....(((.((((...-....)))).)))..)))))))))))........ ( -33.90, z-score = -2.58, R) >droEre2.scaffold_4784 2377787 113 - 25762168 GGCAAAUGGCUUCGUAGUACUGGUUCUUCUCAGUGCAUGUGUGUAUGUCCUGUACUCACGCGUUUUCCAGGCGCCAAAACCUGAUGGCUCCUCUGACUGGGAAACAAUGAAAC .((.............(((((((......)))))))..(((.((((.....)))).)))))(((((((((..((((........))))((....)))))))))))........ ( -33.20, z-score = -1.34, R) >droSim1.chr3L 1925087 113 - 22553184 AGCAUAUGGCUGGAUAGCACUAAUUUCUCUCAGUGCAUGUGUGUAUGUCCGCCAUGCACGCGUUUUCCAGGCGCCAAAAACUGAUGGCCCCUCCGACUGGAAAACAAUGAAAC (((.....))).....(((((..........)))))..(((((((((.....)))))))))((((((((((.((((........)))).))......))))))))........ ( -35.10, z-score = -1.86, R) >consensus _______GGCUGCAUAGCACUGGUUUCUCUCAGUGCAUGUGUGUAUGUCCUGCAUGCACGCGUUUUCCAGGCGCCAAAAACUGAUGGCCCCUCCGACUGGAAAACAAUGAAAC ................(((((((......)))))))..(((((((((.....)))))))))((((((((((.((((........)))).))......))))))))........ (-34.08 = -34.32 + 0.24)

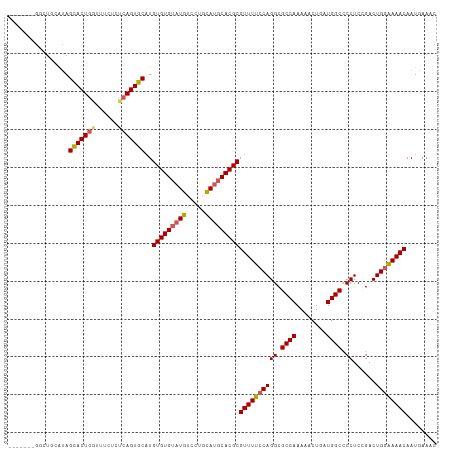
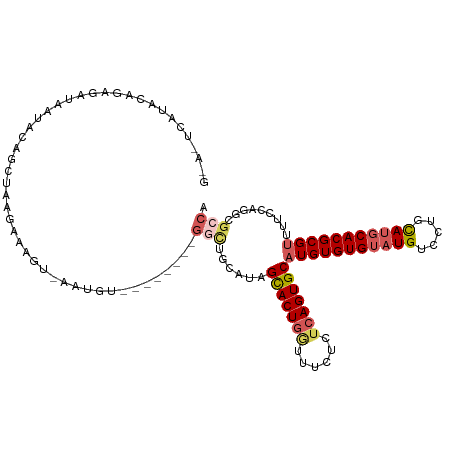
| Location | 2,362,886 – 2,362,986 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 71.59 |
| Shannon entropy | 0.49336 |
| G+C content | 0.44842 |
| Mean single sequence MFE | -29.98 |
| Consensus MFE | -20.12 |
| Energy contribution | -21.28 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.805341 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 2362886 100 - 24543557 -------UUCAGAGAUAAUACAGCUAAGAAAGG-AAUAU--------GGCUGCAUAGCACUGUUUUCUCUCAGUGCCUGUGUGUAUGUCCUGCAUGCACGCGUUUUCCAGGCGCCA -------...(((((.((((..((((.(...((-.....--------..)).).))))..)))).)))))..(((((((((((((((.....)))))))))(.....))))))).. ( -30.60, z-score = -1.11, R) >droSec1.super_2 2387669 100 - 7591821 GCAAUCAUUCAGAGAUAAUACAGAUAUGAAAUU-AAUGU---------------UAGCACUGGUUUCUCUCAGUGCAUGUGUGUAUGUCCGCCAUGCACGCGUUUUCCAGGCGCCA ...........((((............((((((-(.((.---------------...)).))))))))))).(((((((.(((......))))))))))(((((.....))))).. ( -26.10, z-score = -0.92, R) >droYak2.chr3L 14623244 101 + 24197627 GGAAUCAUAUAUGGAUAAUAUAAGUAAAUAUG---------------GUUUUAGUAGCACUGGUUCUUCUCAGUGCAUGUGUGUAUGUCCUGUAUGCACGCGUUUUCCAGGCGCCA ((..((.(((((.....)))))........((---------------(........(((((((......)))))))(((((((((((.....)))))))))))...)))))..)). ( -28.10, z-score = -1.48, R) >droEre2.scaffold_4784 2377824 116 - 25762168 GGAGUCAUAUGGGGAUCAUACAGGUAAAAAAGCGAGUGUUGGCAAAUGGCUUCGUAGUACUGGUUCUUCUCAGUGCAUGUGUGUAUGUCCUGUACUCACGCGUUUUCCAGGCGCCA .((((.(((.(((.((((((((.........(((((.((((.....))))))))).(((((((......))))))).)))))).)).))))))))))..(((((.....))))).. ( -34.10, z-score = -0.72, R) >droSim1.chr3L 1925124 106 - 22553184 ---------CAGUGAUAAUACAGCUAUGAAAUU-AAUGUUAGCAUAUGGCUGGAUAGCACUAAUUUCUCUCAGUGCAUGUGUGUAUGUCCGCCAUGCACGCGUUUUCCAGGCGCCA ---------.............((((.......-.....))))....(.(((((..(((((..........)))))(((((((((((.....)))))))))))..))))).).... ( -31.00, z-score = -0.96, R) >consensus G_A_UCAUACAGAGAUAAUACAGCUAAGAAAGU_AAUGU________GGCUGCAUAGCACUGGUUUCUCUCAGUGCAUGUGUGUAUGUCCUGCAUGCACGCGUUUUCCAGGCGCCA ...............................................(((......(((((((......)))))))(((((((((((.....))))))))))).........))). (-20.12 = -21.28 + 1.16)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:57:31 2011