
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 2,346,258 – 2,346,350 |
| Length | 92 |
| Max. P | 0.997291 |

| Location | 2,346,258 – 2,346,350 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | forward |
| Mean pairwise identity | 70.14 |
| Shannon entropy | 0.57537 |
| G+C content | 0.41863 |
| Mean single sequence MFE | -26.62 |
| Consensus MFE | -12.14 |
| Energy contribution | -12.95 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.35 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.07 |
| SVM RNA-class probability | 0.997291 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
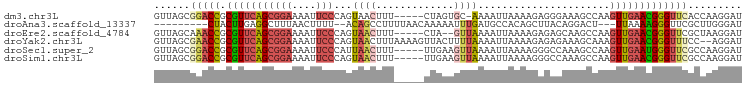
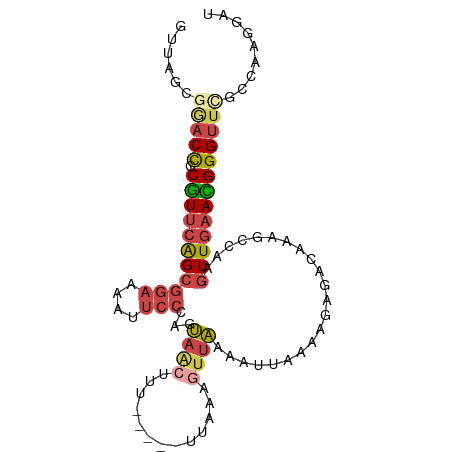
>dm3.chr3L 2346258 92 + 24543557 GUUAGCGGACCGCGUUCAGCGGAAAAUUCCCAGUAACUUU-----CUAGUGC-AAAAUUAAAAGAGGGAAAGCCAAGUUGAACGGGUUCACCAAGGAU ......(((((.((((((((((....(((((.....((((-----.((((..-...)))))))).)))))..))..)))))))))))))......... ( -26.80, z-score = -2.29, R) >droAna3.scaffold_13337 394404 84 + 23293914 ---------CUACUUGAGGCUUUAACUUUU--ACAGCCUUUUAACAAAAAUUUGAUGCCACAGCUUACAGGACU---UUAAAAGGGUUCGCUUGGGAU ---------......((((((.((.....)--).))))))..............((.(((.(((.....(((((---(.....)))))))))))).)) ( -14.90, z-score = -0.03, R) >droEre2.scaffold_4784 2359928 91 + 25762168 GUUAGCAAACCGCGUUCAGCGGAAAAUUCCCAGUAACUUU-----CUA--GUUAAAAUUAAAAGAGAGCAAGCCAAGUUGAACGGGUUCGCUAAGGAU .(((((.((((.(((((((((((....)))......((((-----((.--.(((....))).))))))........)))))))))))).))))).... ( -28.10, z-score = -3.80, R) >droYak2.chr3L 14606296 96 - 24197627 GUUAGCGAACCGCGUUCAGCGGAAAAUUCCCAGUAACUUUAAAAGUUACUUUUAAAAUUAAAAGAGAGAAAGCAAAGUUGAACGGGUUUCC--AGGAU ....(.(((((.(((((((((((....))).(((((((.....)))))))..........................))))))))))))).)--..... ( -25.30, z-score = -2.95, R) >droSec1.super_2 2371156 93 + 7591821 GUUAGCGGACCGCGUUCAGCGGAAAAUUCCCAUUAACUUU-----UUGAAGUUAAAAUUAAAAGGGCCAAAGCCAAGUUGAAUGGGUUCGCCAAGGAU ....(((((((.(((((((((((....)))..(((((((.-----...))))))).........(((....)))..)))))))))))))))....... ( -31.90, z-score = -3.56, R) >droSim1.chr3L 1907891 93 + 22553184 GUUAGCGGACCGCGUUCAGCGGAAAAUUCCCAGUAACUUU-----UUGAAGUUAAAAUUAAAAGGGCCAAAGCCAAGUUGAACGGGUUCGCCAAGGAU ....(((((((.(((((((((((....)))...((((((.-----...))))))..........(((....)))..)))))))))))))))....... ( -32.70, z-score = -3.71, R) >consensus GUUAGCGGACCGCGUUCAGCGGAAAAUUCCCAGUAACUUU_____UUAAAGUUAAAAUUAAAAGAGACAAAGCCAAGUUGAACGGGUUCGCCAAGGAU ......(((((.(((((((((((....)))......((((....................))))............)))))))))))))......... (-12.14 = -12.95 + 0.81)
| Location | 2,346,258 – 2,346,350 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 70.14 |
| Shannon entropy | 0.57537 |
| G+C content | 0.41863 |
| Mean single sequence MFE | -23.70 |
| Consensus MFE | -10.58 |
| Energy contribution | -13.42 |
| Covariance contribution | 2.84 |
| Combinations/Pair | 1.32 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.993507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
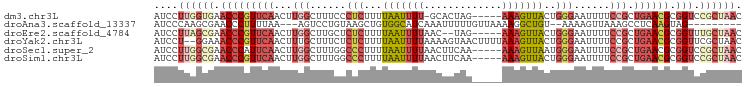
>dm3.chr3L 2346258 92 - 24543557 AUCCUUGGUGAACCCGUUCAACUUGGCUUUCCCUCUUUUAAUUUU-GCACUAG-----AAAGUUACUGGGAAUUUUCCGCUGAACGCGGUCCGCUAAC ....((((((.(((((((((...(((..(((((.....(((((((-.......-----)))))))..)))))....))).)))))).))).)))))). ( -26.50, z-score = -2.94, R) >droAna3.scaffold_13337 394404 84 - 23293914 AUCCCAAGCGAACCCUUUUAA---AGUCCUGUAAGCUGUGGCAUCAAAUUUUUGUUAAAAGGCUGU--AAAAGUUAAAGCCUCAAGUAG--------- .......((.....((((((.---.........((((.(((((.........)))))...)))).)--))))).....)).........--------- ( -10.80, z-score = 0.72, R) >droEre2.scaffold_4784 2359928 91 - 25762168 AUCCUUAGCGAACCCGUUCAACUUGGCUUGCUCUCUUUUAAUUUUAAC--UAG-----AAAGUUACUGGGAAUUUUCCGCUGAACGCGGUUUGCUAAC ....((((((((((((((((...(((.....((((...(((((((...--...-----)))))))..)))).....))).)))))).)))))))))). ( -28.70, z-score = -4.36, R) >droYak2.chr3L 14606296 96 + 24197627 AUCCU--GGAAACCCGUUCAACUUUGCUUUCUCUCUUUUAAUUUUAAAAGUAACUUUUAAAGUUACUGGGAAUUUUCCGCUGAACGCGGUUCGCUAAC ....(--((.((((((((((........(((((.....((((((((((((...))))))))))))..)))))........)))))).))))..))).. ( -22.89, z-score = -2.88, R) >droSec1.super_2 2371156 93 - 7591821 AUCCUUGGCGAACCCAUUCAACUUGGCUUUGGCCCUUUUAAUUUUAACUUCAA-----AAAGUUAAUGGGAAUUUUCCGCUGAACGCGGUCCGCUAAC ....((((((..((((........(((....))).........(((((((...-----.)))))))))))......((((.....))))..)))))). ( -25.30, z-score = -2.39, R) >droSim1.chr3L 1907891 93 - 22553184 AUCCUUGGCGAACCCGUUCAACUUGGCUUUGGCCCUUUUAAUUUUAACUUCAA-----AAAGUUACUGGGAAUUUUCCGCUGAACGCGGUCCGCUAAC ....((((((.(((((((((...(((......(((...(((((((........-----)))))))..)))......))).)))))).))).)))))). ( -28.00, z-score = -3.07, R) >consensus AUCCUUGGCGAACCCGUUCAACUUGGCUUUCUCUCUUUUAAUUUUAACUUUAA_____AAAGUUACUGGGAAUUUUCCGCUGAACGCGGUCCGCUAAC ....((((((.(((((((((...(((......(((...(((((((.............)))))))..)))......))).)))))).))).)))))). (-10.58 = -13.42 + 2.84)

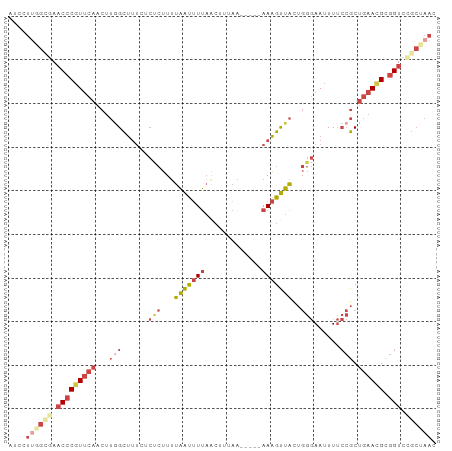
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:57:28 2011