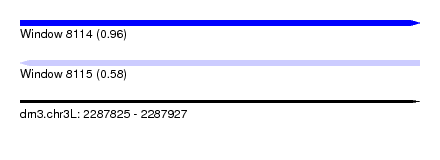
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 2,287,825 – 2,287,927 |
| Length | 102 |
| Max. P | 0.964742 |

| Location | 2,287,825 – 2,287,927 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 80.00 |
| Shannon entropy | 0.36325 |
| G+C content | 0.53019 |
| Mean single sequence MFE | -33.06 |
| Consensus MFE | -22.46 |
| Energy contribution | -22.82 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.964742 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
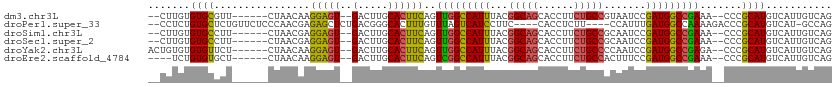
>dm3.chr3L 2287825 102 + 24543557 --CUUGUGUGCGUU------CUAACAAGGAGU--GACUUGCACUUCAGUUGGCCAUUUACGGCAGCACCUUCUGCCGUAAUCCGAUGGCCGAAA--CCCGCAUGUCAUUGUCAG --(((((.......------...))))).(((--(((.(((.......(((((((((((((((((......))))))))....)))))))))..--...))).))))))..... ( -38.40, z-score = -3.63, R) >droPer1.super_33 233769 103 + 967471 --CCUCUGUGCUCUGUUCUCCCAACGAGAGCUCUGACGGGCACUUUGUUUAGUCAUCCUUC----CACCUCUU----CCAUUUGAUGGCCAAAAGACCCGCAUGUCAU-GCCAG --...(((.((...((((((.....))))))..((.((((..((((.....((((((....----........----......))))))..)))).))))))......-))))) ( -22.97, z-score = -0.50, R) >droSim1.chr3L 1849855 102 + 22553184 --CUUGUGUGCCUU------CUAACGAGGAGU--GACUUGCACUUCAGUUGGCCAUUUACGGCAGCACCUUCUGCCGCAAUCCGAUGGCCGAAA--CCCGCAUGUCAUUGUCAG --.........(((------(....))))(((--(((.(((.......(((((((((..((((((......))))))......)))))))))..--...))).))))))..... ( -35.30, z-score = -2.48, R) >droSec1.super_2 2313207 102 + 7591821 --CUUGUGUGCCUU------CUAACGAGGAGU--GACUUGCACUUCAGUUGGCCAUUUACGGCAGCACCUUCUGCCGCAAUCCGAUGGCCGAAA--CCCGCAUGUCAUUGUCAG --.........(((------(....))))(((--(((.(((.......(((((((((..((((((......))))))......)))))))))..--...))).))))))..... ( -35.30, z-score = -2.48, R) >droYak2.chr3L 14546426 104 - 24197627 ACUGUGUGUGUUCU------CUAACAAGGAGU--GACUUGCACUUCAGUUGGCCAUUUACGGCAGCACCUUCUGCCCCAAUCCGAUGGCCGAGA--CCCGCAUGUCAUUGUCAG ...(((((.(.(((------(......(((((--(.....))))))....(((((((...(((((......))))).......)))))))))))--.))))))........... ( -33.80, z-score = -1.87, R) >droEre2.scaffold_4784 2299455 100 + 25762168 ----UCUGUGUGCU------CUAACAAGGAGU--GACUUGCACUUCAGUCGGCCAUUUACGGCAGCACCUUCUGCCACUUUCCGAUGGCCGAAA--CCCGCAUGUCAUUGUCAG ----(((.(((...------...))).)))((--(((.(((.......(((((((((...(((((......))))).......)))))))))..--...))).)))))...... ( -32.60, z-score = -2.26, R) >consensus __CUUGUGUGCCCU______CUAACAAGGAGU__GACUUGCACUUCAGUUGGCCAUUUACGGCAGCACCUUCUGCCGCAAUCCGAUGGCCGAAA__CCCGCAUGUCAUUGUCAG ..................................(((.(((.......(((((((((...(((((......))))).......))))))))).......))).)))........ (-22.46 = -22.82 + 0.36)
| Location | 2,287,825 – 2,287,927 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 80.00 |
| Shannon entropy | 0.36325 |
| G+C content | 0.53019 |
| Mean single sequence MFE | -34.83 |
| Consensus MFE | -21.62 |
| Energy contribution | -22.82 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.578523 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
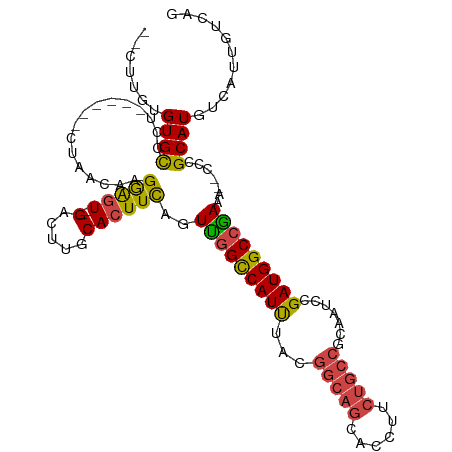
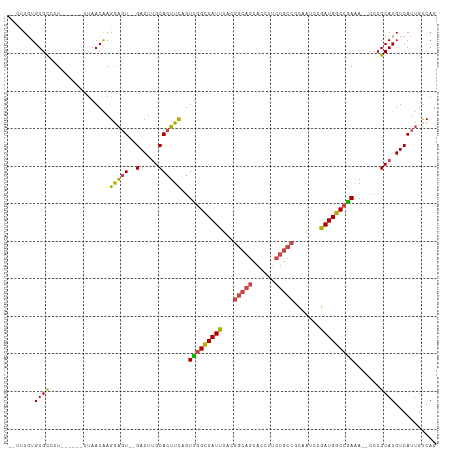
>dm3.chr3L 2287825 102 - 24543557 CUGACAAUGACAUGCGGG--UUUCGGCCAUCGGAUUACGGCAGAAGGUGCUGCCGUAAAUGGCCAACUGAAGUGCAAGUC--ACUCCUUGUUAG------AACGCACACAAG-- (((((((((((.((((..--((..((((((....(((((((((......)))))))))))))))....))..)))).)))--)....)))))))------............-- ( -38.30, z-score = -3.12, R) >droPer1.super_33 233769 103 - 967471 CUGGC-AUGACAUGCGGGUCUUUUGGCCAUCAAAUGG----AAGAGGUG----GAAGGAUGACUAAACAAAGUGCCCGUCAGAGCUCUCGUUGGGAGAACAGAGCACAGAGG-- (((..-.((((..((..(((((((.(((.((......----..))))).----))))))).(((......)))))..))))..(((((.(((.....)))))))).)))...-- ( -29.10, z-score = -0.24, R) >droSim1.chr3L 1849855 102 - 22553184 CUGACAAUGACAUGCGGG--UUUCGGCCAUCGGAUUGCGGCAGAAGGUGCUGCCGUAAAUGGCCAACUGAAGUGCAAGUC--ACUCCUCGUUAG------AAGGCACACAAG-- (((((..((((.((((..--((..((((((....(((((((((......)))))))))))))))....))..)))).)))--)......)))))------............-- ( -34.90, z-score = -1.39, R) >droSec1.super_2 2313207 102 - 7591821 CUGACAAUGACAUGCGGG--UUUCGGCCAUCGGAUUGCGGCAGAAGGUGCUGCCGUAAAUGGCCAACUGAAGUGCAAGUC--ACUCCUCGUUAG------AAGGCACACAAG-- (((((..((((.((((..--((..((((((....(((((((((......)))))))))))))))....))..)))).)))--)......)))))------............-- ( -34.90, z-score = -1.39, R) >droYak2.chr3L 14546426 104 + 24197627 CUGACAAUGACAUGCGGG--UCUCGGCCAUCGGAUUGGGGCAGAAGGUGCUGCCGUAAAUGGCCAACUGAAGUGCAAGUC--ACUCCUUGUUAG------AGAACACACACAGU (((((((((((.((((..--((..((((((....(((.(((((......))))).)))))))))....))..)))).)))--)....)))))))------.............. ( -36.20, z-score = -1.55, R) >droEre2.scaffold_4784 2299455 100 - 25762168 CUGACAAUGACAUGCGGG--UUUCGGCCAUCGGAAAGUGGCAGAAGGUGCUGCCGUAAAUGGCCGACUGAAGUGCAAGUC--ACUCCUUGUUAG------AGCACACAGA---- (((((((((((.((((..--((((((((((..(....((((((......)))))))..))))))))..))..)))).)))--)....)))))))------..........---- ( -35.60, z-score = -1.71, R) >consensus CUGACAAUGACAUGCGGG__UUUCGGCCAUCGGAUUGCGGCAGAAGGUGCUGCCGUAAAUGGCCAACUGAAGUGCAAGUC__ACUCCUCGUUAG______AAAGCACACAAG__ (((((...(((.((((........((((((....(((((((((......)))))))))))))))........)))).))).........))))).................... (-21.62 = -22.82 + 1.20)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:57:23 2011