| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,157,641 – 4,157,719 |
| Length | 78 |
| Max. P | 0.603689 |

| Location | 4,157,641 – 4,157,719 |
|---|---|
| Length | 78 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 66.09 |
| Shannon entropy | 0.60342 |
| G+C content | 0.27591 |
| Mean single sequence MFE | -15.14 |
| Consensus MFE | -4.10 |
| Energy contribution | -4.30 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.53 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.27 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.603689 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
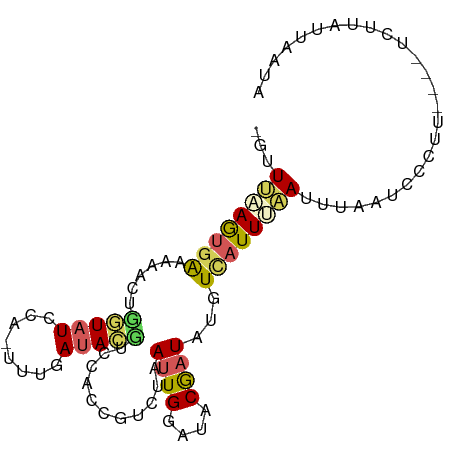
>dm3.chr2L 4157641 78 + 23011544 -GUUUCAGUGAAAAACUGGUAU-----UUGAUACUUC-------UAAUUGGAUACGAUAUGUCAUUUCAUUUAAUCUCUU----UCUUAUUAAUA -....((((.....))))((((-----(..((.....-------..))..)))))((((((......)))...)))....----........... ( -9.90, z-score = -0.44, R) >droEre2.scaffold_4929 4222561 94 + 26641161 -GUUUUAGUGGAAAAGUGAUAUUCAGUUUUAUACGUGCCCCGUCUAAUUGGUUACGAUAUGUUAUUAAAUGUAAAUUAAAAAGGUUUAAUUAAAA -..((((((((((((.((.....)).))))...((((..(((......))).)))).....))))))))....(((((((....))))))).... ( -10.90, z-score = 0.82, R) >droSec1.super_5 2257652 90 + 5866729 -GUUUAAGUGAAAAACUCGUAUCCAAUUUGAUACGUCCACUGUCUAAUUGGAUACGAUAUGUCAUUUAAUUUAAUCCCUU----UUCUAUUAAUA -..((((((((.....((((((((((((.((((.(....))))).))))))))))))....))))))))...........----........... ( -25.10, z-score = -6.14, R) >droSim1.chr2L 4113329 90 + 22036055 -GUUUAAGUGAAAAACUCGUAUCCACUUUGAUACGUCCACUGUCUAAUUGGAUACGAUAUGUCAUUUAAUUUAAUCCCUU----UCUUAUUAAUA -..((((((((.....(((((((((.((.((((.(....))))).)).)))))))))....))))))))...........----........... ( -21.40, z-score = -4.71, R) >droPer1.super_15 326153 82 + 2181545 GCUUACAAGAAAAACACGAUAUC---UUUCACAUCAACUGCGUUAAAAUGAUU-CAAU-UUUCAUUAUAUUUGAUGGCAU--------AUUAAAA ((...............((....---..)).((((((......((.(((((..-....-..))))).)).))))))))..--------....... ( -8.40, z-score = -0.24, R) >consensus _GUUUAAGUGAAAAACUGGUAUCCA_UUUGAUACGUCCACCGUCUAAUUGGAUACGAUAUGUCAUUUAAUUUAAUCCCUU____UCUUAUUAAUA ...((((((((......(((((........)))))...........((((....))))...)))))))).......................... ( -4.10 = -4.30 + 0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:15:33 2011