| Sequence ID | dm3.chr3L |
|---|---|
| Location | 2,273,696 – 2,273,842 |
| Length | 146 |
| Max. P | 0.954302 |

| Location | 2,273,696 – 2,273,842 |
|---|---|
| Length | 146 |
| Sequences | 13 |
| Columns | 165 |
| Reading direction | forward |
| Mean pairwise identity | 68.86 |
| Shannon entropy | 0.66404 |
| G+C content | 0.49917 |
| Mean single sequence MFE | -44.61 |
| Consensus MFE | -24.00 |
| Energy contribution | -24.09 |
| Covariance contribution | 0.09 |
| Combinations/Pair | 1.30 |
| Mean z-score | -0.51 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.679040 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
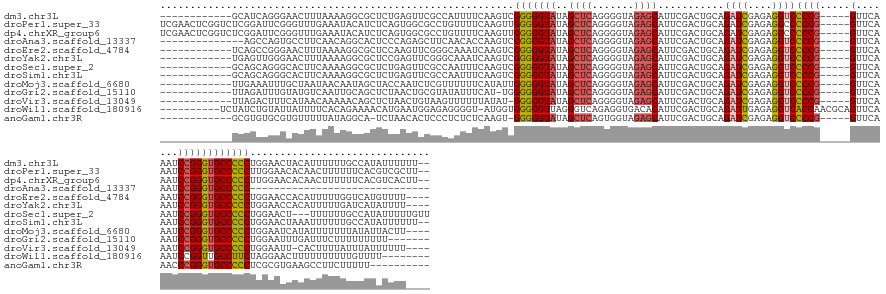
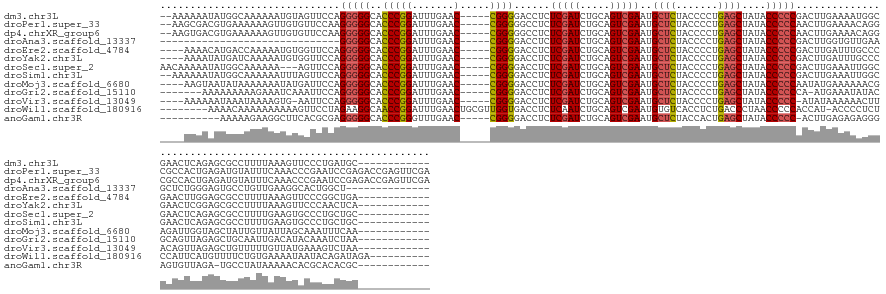
>dm3.chr3L 2273696 146 + 24543557 ------------GCAUCAGGGAACUUUAAAAGGCGCUCUGAGUUCGCCAUUUUCAAGUCGGGGGUAUAGCUCAGGGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCG-----GUUCAAAUCCGGGUGCCCCCUGGAACUACAUUUUUUGCCAUAUUUUUU-- ------------(((((((((..(((....)))..))))))((((((.(((..(.....)..)))...)).((((((....(((......))).((((....))))((((-----(.......)))))...)))))))))).........)))..........-- ( -44.50, z-score = -0.26, R) >droPer1.super_33 215787 158 + 967471 UCGAACUCGGUCUCGGAUUCGGGUUUGAAAUACAUCUCAGUGGCGCCUGUUUUCAAGUUGGGGGUAUAGCUCAGGGGUAGAGCAUUCGACUGCAGAUCGAGAGGCCCCCG-----GUUCAAAUCCGGGUGCCCCCUUGGAACACAACUUUUUUCACGUCGCUU-- ((((((((((........))))))))))......(((.((.((((((((..((..((..((((((...((((.......)))).((((((....).)))))..)))))).-----.))..))..))))))))..)).))).......................-- ( -53.60, z-score = -0.56, R) >dp4.chrXR_group6 12194707 158 + 13314419 UCGAACUCGGUCUCGGAUUCGGGUUUGAAAUACAUCUCAGUGGCGCCUGUUUUCAAGUUGGGGGUAUAGCUCAGGGGUAGAGCAUUCGACUGCAGAUCGAGAGGCCCCCG-----GUUCAAAUCCGGGUGCCCCCUUGGAACACAACUUUUUUCACGUCACUU-- ((((((((((........))))))))))......(((.((.((((((((..((..((..((((((...((((.......)))).((((((....).)))))..)))))).-----.))..))..))))))))..)).))).......................-- ( -53.60, z-score = -0.80, R) >droAna3.scaffold_13337 325441 116 + 23293914 --------------AGCCAGUGCCUUCAACAGGCACUCCCAGAGCUUCAACACCAAGUCGGGGGUAUAGCUCAGGGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCG-----GUUCAAAUCCGGGUGCCCCC------------------------------ --------------.(..(((((((.....)))))))..).(((((...((.((......)).))..))))).((((((..(((......))).((((....))))((((-----(.......))))))))))).------------------------------ ( -45.60, z-score = -2.24, R) >droEre2.scaffold_4784 2283751 144 + 25762168 ------------UCAGCCGGGAACUUUAAAAGGCGCUCCAAGUUCGGGCAAAUCAAGUCGGGGGUAUAGCUCAGGGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCG-----GUUCAAAUCCGGGUGCCCCCUGGAACCACAUUUUUGGUCAUGUUUU---- ------------..(((((((...........((((.((......))))......(((((((......((((.......)))).))))))))).((((....))))))))-----)))....((((((.....))))))((((......))))........---- ( -45.10, z-score = 0.41, R) >droYak2.chr3L 14532289 144 - 24197627 ------------UGAGUUGGGAACUUUAAAAGGCGCUCCGAGUUCGGGCAAAUCAAGUCGGGGGUAUAGCUCAGGGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCG-----GUUCAAAUCCGGGUGCCCCCUGGAACCACAUUUUUGAUCAUAUUUU---- ------------(((((((((...........((.((((((.((.((.....)))).))))))))...((((.......)))).))))))).))((((((((((..((((-----(.......)))))..))...((....))...)))))))).......---- ( -44.00, z-score = 0.40, R) >droSec1.super_2 2299268 145 + 7591821 ------------GCAGCAGGGCACUUCAAAAGGCGCUCUGAGUUCGCCAAUUUCAAGUCGGGGGUAUAGCUCAGGGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCG-----GUUCAAAUCCGGGUGCCCCCUGGAACU---UUUUUUGCCAUAUUUUUGUU ------------((((((((((.((......)).))))))(((((((..((..(......)..))...)).((((((....(((......))).((((....))))((((-----(.......)))))...)))))))))))---....))))............ ( -47.60, z-score = -0.40, R) >droSim1.chr3L 1835847 146 + 22553184 ------------GCAGCAGGGCACUUCAAAAGGCGCUCUGAGUUCGCCAAUUUCAAGUCGGGGGUAUAGCUCAGGGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCG-----GUUCAAAUCCGGGUGCCCCCUGGAACUAAAUUUUUUGCCAUAUUUUUU-- ------------((((((((((.((......)).))))))(((((((..((..(......)..))...)).((((((....(((......))).((((....))))((((-----(.......)))))...))))))))))).......))))..........-- ( -48.20, z-score = -0.85, R) >droMoj3.scaffold_6680 15669885 144 - 24764193 ------------UUGAAAUUUGCUAAUAACAAUAGCUACCAAUCUCGUUUUUUCAUAUUGGGGGUAUAGCUCAGGGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCG-----GUUCAAAUCCGGGUGCCCCCUGGAAUCAUAUUUUUUUAUAUUACUU---- ------------.(((.....((((.......))))..(((.....(......).....(((((((..((((.......))))...........((((....))))((((-----(.......)))))))))))))))..)))..................---- ( -36.40, z-score = -0.13, R) >droGri2.scaffold_15110 11188050 140 + 24565398 ------------UUAGAUUUGUAUGUCAAUUGCAGCUCUAACUGCGUAUAUUUCAU-UGGGGGGUAUAGCUCAGGGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCG-----GUUCAAAUCCGGGUGCCCCCUGGAAUUUGAUUUCUUUUUUUUU------- ------------...((((((((.((((((....((((((.(((.((((((..(..-...)..)))).)).)))...))))))))).)))))))))))((((((((((((-----(.......)))))..((....)).....)))))))).......------- ( -44.50, z-score = -1.43, R) >droVir3.scaffold_13049 11731604 142 + 25233164 ------------UUAGACUUUCAUAACAAAAACAGCUCUAACUGUAAGUUUUUUAUAU-GGGGGUAUAGCUCAGGGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCG-----GUUCAAAUCCGGGUGCCCCCUGGAAUU-CACUUUUAUUUAUUUUUU---- ------------...((.(((((........((((......)))).............-(((((((..((((.......))))...........((((....))))((((-----(.......))))))))))))))))).)-).................---- ( -38.90, z-score = -1.11, R) >droWil1.scaffold_180916 1734078 146 + 2700594 ----------UCUAUCUGUAUUAUUUUCACAGAAAACAUGAAUGGAGAGGGGU-AUGGUGGGGGUUAGGGUCAGAGGUGACACAUUCGACUGCAGAUUGAGAGGUCACCAACGCAGUUCAAAUCCGGUUGCCUUCUAGGAACUUUUUUUUUUGUUUU-------- ----------....(((((.........)))))(((((.....((((((((((-.....((((((..((((..((((((((.(.((((((....).))))).)))))))........))..))))....)))))).....)))))))))).))))).-------- ( -33.00, z-score = 1.16, R) >anoGam1.chr3R 22015911 136 - 53272125 ------------GCGUGUGCGUGUUUUUAUAGGCA-UCUAACACUCCCUCUCUCAAGU-GGGGGUAUAGCUCAGUGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCG-----GUUCAAACCCGGGUGCCCCCUCGCGUGAAGCCUUCUUUUU---------- ------------((((((..((((((....)))))-)...))))........(((.((-(((((....((((.......)))).((((((....).))))).((..((((-----(.......)))))..))))))))).))).)).........---------- ( -44.90, z-score = -0.81, R) >consensus ____________UCAGAUGGGGAUUUUAAAAGGCGCUCUGAGUUCGCCAAUUUCAAGUCGGGGGUAUAGCUCAGGGGUAGAGCAUUCGACUGCAGAUCGAGAGGUCCCCG_____GUUCAAAUCCGGGUGCCCCCUGGAACCACAUUUUUUUUCAUAUUUU____ ...........................................................((((((...((((.(((...((((.......((..((((....))))..)).....))))...))))))))))))).............................. (-24.00 = -24.09 + 0.09)
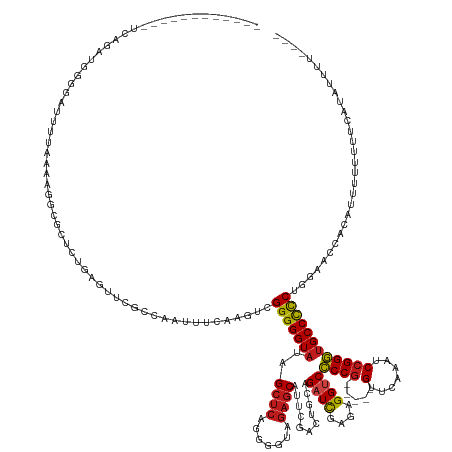
| Location | 2,273,696 – 2,273,842 |
|---|---|
| Length | 146 |
| Sequences | 13 |
| Columns | 165 |
| Reading direction | reverse |
| Mean pairwise identity | 68.86 |
| Shannon entropy | 0.66404 |
| G+C content | 0.49917 |
| Mean single sequence MFE | -41.69 |
| Consensus MFE | -21.75 |
| Energy contribution | -21.86 |
| Covariance contribution | 0.10 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.20 |
| Structure conservation index | 0.52 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.60 |
| SVM RNA-class probability | 0.954302 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
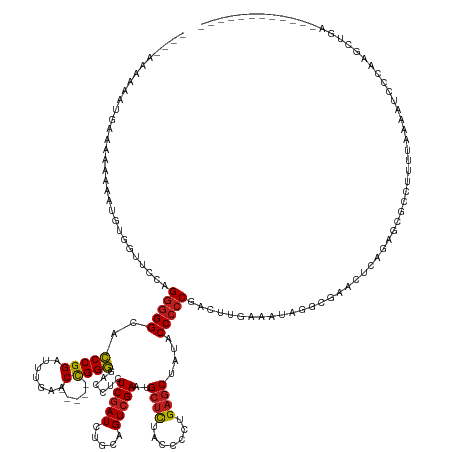
>dm3.chr3L 2273696 146 - 24543557 --AAAAAAUAUGGCAAAAAAUGUAGUUCCAGGGGGCACCCGGAUUUGAAC-----CGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCCCUGAGCUAUACCCCCGACUUGAAAAUGGCGAACUCAGAGCGCCUUUUAAAGUUCCCUGAUGC------------ --..........(((.....((((((((.(((((...(((((.......)-----))))((.(..(((((.....)))))..).))..))))))))))))).....(((((.((((.((((.........)))))))).)))))......)))------------ ( -44.30, z-score = -1.64, R) >droPer1.super_33 215787 158 - 967471 --AAGCGACGUGAAAAAAGUUGUGUUCCAAGGGGGCACCCGGAUUUGAAC-----CGGGGGCCUCUCGAUCUGCAGUCGAAUGCUCUACCCCUGAGCUAUACCCCCAACUUGAAAACAGGCGCCACUGAGAUGUAUUUCAAACCCGAAUCCGAGACCGAGUUCGA --..(((((.........))))).......((((((.((.(..((..(..-----.(((((....(((((.....)))))..((((.......))))....)))))...)..))..).)).)))..(((((....)))))..)))(((..((....))..))).. ( -46.10, z-score = -0.16, R) >dp4.chrXR_group6 12194707 158 - 13314419 --AAGUGACGUGAAAAAAGUUGUGUUCCAAGGGGGCACCCGGAUUUGAAC-----CGGGGGCCUCUCGAUCUGCAGUCGAAUGCUCUACCCCUGAGCUAUACCCCCAACUUGAAAACAGGCGCCACUGAGAUGUAUUUCAAACCCGAAUCCGAGACCGAGUUCGA --..(..(((..(......)..)))..)..((((((.((.(..((..(..-----.(((((....(((((.....)))))..((((.......))))....)))))...)..))..).)).)))..(((((....)))))..)))(((..((....))..))).. ( -46.20, z-score = -0.28, R) >droAna3.scaffold_13337 325441 116 - 23293914 ------------------------------GGGGGCACCCGGAUUUGAAC-----CGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCCCUGAGCUAUACCCCCGACUUGGUGUUGAAGCUCUGGGAGUGCCUGUUGAAGGCACUGGCU-------------- ------------------------------.((((..(((((.......)-----))))..))))(((((.....)))))..(((....(((.((((((((((........)))))...))))).)))(((((((.....)))))))))).-------------- ( -50.00, z-score = -2.66, R) >droEre2.scaffold_4784 2283751 144 - 25762168 ----AAAACAUGACCAAAAAUGUGGUUCCAGGGGGCACCCGGAUUUGAAC-----CGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCCCUGAGCUAUACCCCCGACUUGAUUUGCCCGAACUUGGAGCGCCUUUUAAAGUUCCCGGCUGA------------ ----...((((........)))).((((((((((((((((((.......)-----))))........((((...(((((...((((.......))))........))))).)))))))))...))))))))(((.............)))...------------ ( -44.32, z-score = -0.97, R) >droYak2.chr3L 14532289 144 + 24197627 ----AAAAUAUGAUCAAAAAUGUGGUUCCAGGGGGCACCCGGAUUUGAAC-----CGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCCCUGAGCUAUACCCCCGACUUGAUUUGCCCGAACUCGGAGCGCCUUUUAAAGUUCCCAACUCA------------ ----.......((((((...((..(.((.(((((...(((((.......)-----))))..))))).)).)..))((((...((((.......))))........)))))))))).(((((....))).))......................------------ ( -42.00, z-score = -0.97, R) >droSec1.super_2 2299268 145 - 7591821 AACAAAAAUAUGGCAAAAAA---AGUUCCAGGGGGCACCCGGAUUUGAAC-----CGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCCCUGAGCUAUACCCCCGACUUGAAAUUGGCGAACUCAGAGCGCCUUUUGAAGUGCCCUGCUGC------------ ...........(((......---(((((.(((((...(((((.......)-----))))((.(..(((((.....)))))..).))..)))))))))).........((((.(((..((((.........)))).))).))))))).......------------ ( -44.20, z-score = -1.28, R) >droSim1.chr3L 1835847 146 - 22553184 --AAAAAAUAUGGCAAAAAAUUUAGUUCCAGGGGGCACCCGGAUUUGAAC-----CGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCCCUGAGCUAUACCCCCGACUUGAAAUUGGCGAACUCAGAGCGCCUUUUGAAGUGCCCUGCUGC------------ --.........(((........((((((.(((((...(((((.......)-----))))((.(..(((((.....)))))..).))..)))))))))))........((((.(((..((((.........)))).))).))))))).......------------ ( -45.30, z-score = -1.45, R) >droMoj3.scaffold_6680 15669885 144 + 24764193 ----AAGUAAUAUAAAAAAAUAUGAUUCCAGGGGGCACCCGGAUUUGAAC-----CGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCCCUGAGCUAUACCCCCAAUAUGAAAAAACGAGAUUGGUAGCUAUUGUUAUUAGCAAAUUUCAA------------ ----..........................(((((..(((((.......)-----))))......(((((.....)))))..((((.......))))....))))).............((((((....((((.......)))).))))))..------------ ( -36.50, z-score = -1.15, R) >droGri2.scaffold_15110 11188050 140 - 24565398 -------AAAAAAAAAGAAAUCAAAUUCCAGGGGGCACCCGGAUUUGAAC-----CGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCCCUGAGCUAUACCCCCCA-AUGAAAUAUACGCAGUUAGAGCUGCAAUUGACAUACAAAUCUAA------------ -------.............((((......(((((..(((((.......)-----))))......(((((.....)))))..((((.......)))).....))))).-...........(((((....)))))..)))).............------------ ( -37.70, z-score = -2.30, R) >droVir3.scaffold_13049 11731604 142 - 25233164 ----AAAAAAUAAAUAAAAGUG-AAUUCCAGGGGGCACCCGGAUUUGAAC-----CGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCCCUGAGCUAUACCCCC-AUAUAAAAAACUUACAGUUAGAGCUGUUUUUGUUAUGAAAGUCUAA------------ ----........(((((((...-.......(((((..(((((.......)-----))))......(((((.....)))))..((((.......))))....)))))-.............(((((....))))))))))))............------------ ( -35.20, z-score = -1.18, R) >droWil1.scaffold_180916 1734078 146 - 2700594 --------AAAACAAAAAAAAAAGUUCCUAGAAGGCAACCGGAUUUGAACUGCGUUGGUGACCUCUCAAUCUGCAGUCGAAUGUGUCACCUCUGACCCUAACCCCCACCAU-ACCCCUCUCCAUUCAUGUUUUCUGUGAAAAUAAUACAGAUAGA---------- --------................(((.(((((((((...(.((((((.(((((....(((....)))...))))))))))).)((((....))))...............-...............))))))))).)))...............---------- ( -24.40, z-score = -0.05, R) >anoGam1.chr3R 22015911 136 + 53272125 ----------AAAAAGAAGGCUUCACGCGAGGGGGCACCCGGGUUUGAAC-----CGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCACUGAGCUAUACCCCC-ACUUGAGAGAGGGAGUGUUAGA-UGCCUAUAAAAACACGCACACGC------------ ----------.......((((......(((((((...(((((.......)-----))))..)))))))(((((((.((....((((.......)))).........-.(((....))).)).)).))))-)))))..................------------ ( -45.70, z-score = -1.53, R) >consensus ____AAAAAAUGAAAAAAAAUGUGGUUCCAGGGGGCACCCGGAUUUGAAC_____CGGGGACCUCUCGAUCUGCAGUCGAAUGCUCUACCCCUGAGCUAUACCCCCGACUUGAAAUAGGCGAACUCAGAGCGCCUUUUAAAAUCCCAAGCUGA____________ ..............................(((((..((((..............))))......(((((.....)))))..((((.......))))....)))))........................................................... (-21.75 = -21.86 + 0.10)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:57:22 2011