
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 2,273,451 – 2,273,542 |
| Length | 91 |
| Max. P | 0.739770 |

| Location | 2,273,451 – 2,273,542 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 82.40 |
| Shannon entropy | 0.30578 |
| G+C content | 0.51884 |
| Mean single sequence MFE | -29.68 |
| Consensus MFE | -19.44 |
| Energy contribution | -20.20 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.65 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.739770 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
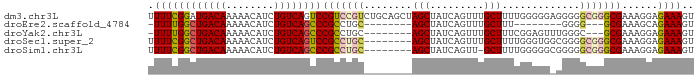
>dm3.chr3L 2273451 91 + 24543557 UUUUCGGAUGACAAAAACAUCUGUCAGUCCGUCCGUCUGCAGCUAGCUAUCAGUUUGCUUUUGGGGGAGGGGGCGGGCGAAAGGAGAAAGU (((((...(((((........)))))((((((((.(((.(((..(((.........))).))).)))...)))))))).....)))))... ( -29.10, z-score = -1.13, R) >droEre2.scaffold_4784 2283523 71 + 25762168 -UUUUGGCUGACAAAAACAUCUGUCAGCCCGCCUGC--------AGCUAUCAGUUUGCUUU--------GGGG---GCGAAAGCAGAAAGU -....((((((((........))))))))(((((.(--------(((.........))..)--------).))---)))............ ( -24.70, z-score = -1.62, R) >droYak2.chr3L 14532047 79 - 24197627 -UUUUGGCUGACAAAAACAUCUGUCAGCCCGCCUGC--------AGCUAUCAGUUUGCUUUCGGAGUUUGGGC---GCGAAAGGAGAAAGU -....((((((((........))))))))((((((.--------((((...((....)).....)))))))))---)(....)........ ( -23.80, z-score = -0.52, R) >droSec1.super_2 2299010 83 + 7591821 UUUUCGGCUGACAAAAACAUCUGUCAGUCCGCCUGC--------AGCUAUCAGUUUGCUUUUGGGUGGCGGGGCGGGCGAAAGGAGAAAGU (((((((((((((........))))))))(((((((--------.((((((............))))))...)))))))....)))))... ( -34.50, z-score = -3.18, R) >droSim1.chr3L 1835591 82 + 22553184 UUUUCGGCUGACAAAAACAUCUGUCAGCCCGCCUGC--------AGCUAUCAGUU-GCUUUUGGGGGCGGGGGCGGGCGAAAGGAGAAAGU (((((((((((((........))))))))(((((((--------.(((..(((..-....)))..)))....)))))))....)))))... ( -36.30, z-score = -3.17, R) >consensus UUUUCGGCUGACAAAAACAUCUGUCAGCCCGCCUGC________AGCUAUCAGUUUGCUUUUGGGGG_GGGGGCGGGCGAAAGGAGAAAGU .((((((((((((........))))))))(((((((........(((.........))).............)))))))......)))).. (-19.44 = -20.20 + 0.76)


| Location | 2,273,451 – 2,273,542 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 82.40 |
| Shannon entropy | 0.30578 |
| G+C content | 0.51884 |
| Mean single sequence MFE | -23.63 |
| Consensus MFE | -13.97 |
| Energy contribution | -15.17 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.609640 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
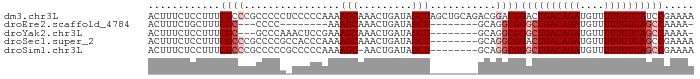
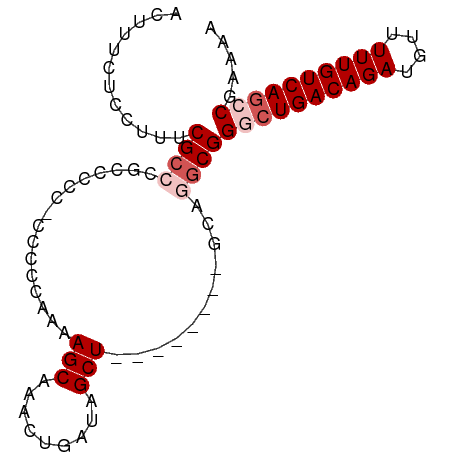
>dm3.chr3L 2273451 91 - 24543557 ACUUUCUCCUUUCGCCCGCCCCCUCCCCCAAAAGCAAACUGAUAGCUAGCUGCAGACGGACGGACUGACAGAUGUUUUUGUCAUCCGAAAA ......(((........((..............))...(((.(((....))))))..)))((((.(((((((....))))))))))).... ( -19.84, z-score = -1.34, R) >droEre2.scaffold_4784 2283523 71 - 25762168 ACUUUCUGCUUUCGC---CCCC--------AAAGCAAACUGAUAGCU--------GCAGGCGGGCUGACAGAUGUUUUUGUCAGCCAAAA- ............(((---(...--------..(((.........)))--------...))))((((((((((....))))))))))....- ( -24.90, z-score = -2.38, R) >droYak2.chr3L 14532047 79 + 24197627 ACUUUCUCCUUUCGC---GCCCAAACUCCGAAAGCAAACUGAUAGCU--------GCAGGCGGGCUGACAGAUGUUUUUGUCAGCCAAAA- ............(((---..........(....)....(((......--------.))))))((((((((((....))))))))))....- ( -22.70, z-score = -1.32, R) >droSec1.super_2 2299010 83 - 7591821 ACUUUCUCCUUUCGCCCGCCCCGCCACCCAAAAGCAAACUGAUAGCU--------GCAGGCGGACUGACAGAUGUUUUUGUCAGCCGAAAA .........(((((.(((((..((........(((.........)))--------)).))))).((((((((....)))))))).))))). ( -23.20, z-score = -2.16, R) >droSim1.chr3L 1835591 82 - 22553184 ACUUUCUCCUUUCGCCCGCCCCCGCCCCCAAAAGC-AACUGAUAGCU--------GCAGGCGGGCUGACAGAUGUUUUUGUCAGCCGAAAA .........(((((.......(((((......(((-........)))--------...)))))(((((((((....)))))))))))))). ( -27.50, z-score = -2.78, R) >consensus ACUUUCUCCUUUCGCCCGCCCCC_CCCCCAAAAGCAAACUGAUAGCU________GCAGGCGGGCUGACAGAUGUUUUUGUCAGCCGAAAA ............((((................(((.........)))...........))))((((((((((....))))))))))..... (-13.97 = -15.17 + 1.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:57:20 2011