| Sequence ID | dm3.chr3L |
|---|---|
| Location | 2,197,518 – 2,197,657 |
| Length | 139 |
| Max. P | 0.686563 |

| Location | 2,197,518 – 2,197,623 |
|---|---|
| Length | 105 |
| Sequences | 11 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 80.37 |
| Shannon entropy | 0.40334 |
| G+C content | 0.50789 |
| Mean single sequence MFE | -31.48 |
| Consensus MFE | -14.56 |
| Energy contribution | -13.78 |
| Covariance contribution | -0.78 |
| Combinations/Pair | 1.45 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.597358 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
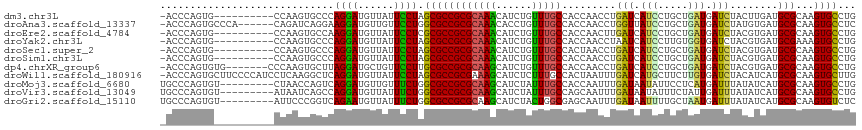
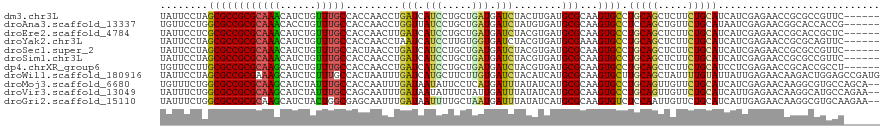
>dm3.chr3L 2197518 105 + 24543557 -ACCCAGUG----------CCAAGUGCCCAGGAUGUUAUUCCUAGCGCCGCGCAAACAUCUGUUUGCCACCAACCUGAUCAUCCUGCUGAUGAUCUACUUGAUGCGCAAGUGCCUG -...(((.(----------((..((((..((((......)))).))))(((((((((....))))))...(((...(((((((.....)))))))...)))..)))...).))))) ( -33.00, z-score = -3.22, R) >droAna3.scaffold_13337 22653722 109 - 23293914 -ACCCAGUGCCCA------CAGAUCAGGAAGGAUGUUGUUCCUGGCGCCGCGCAAACACCUGUUUGCCACCAACCUGGUUAUCCUGCUGAUGAUCUAUGUGAUGCGCAAGUGCCUC -.....((((.((------(((((((...((((((.....((.((......((((((....))))))......)).)).)))))).....)))))..))))..))))......... ( -31.40, z-score = -0.37, R) >droEre2.scaffold_4784 2192393 105 + 25762168 -ACCCAGUG----------CCAAGUGCCAAGGAUGUUAUUCCUCGCGCCGCGCAAACAUCUGUUUGCCACCAACUUGAUCAUCCUGCUGAUGAUCUACGUGAUGCGCAAGUGCCUG -...(((.(----------((..((((..((((......)))).))))(((((((((....))))))(((......(((((((.....)))))))...)))..)))...).))))) ( -32.60, z-score = -2.49, R) >droYak2.chr3L 2150940 105 + 24197627 -ACCCAGUG----------CCAAGUGCCCAGGAUGUUAUUCCUAGCGCCGCGCAAACAUCUGUUUGCCACCAACCUAAUCAUCCUUGUGGUGAUCUACGUGAUGCGAAAGUGCCUG -...(((.(----------((..((((..((((......)))).))))(((((((((....))))))(((.......((((((.....))))))....)))..)))...).))))) ( -29.20, z-score = -1.90, R) >droSec1.super_2 2223535 105 + 7591821 -ACCCAGUG----------CCAAGUGCCCAGGAUGUUAUUCCUAGCGCCGCGCAAACAUCUGUUUGCCACUAACCUGAUCAUCCUGCUGAUGAUCUACGUGAUGCGCAAGUGCCUG -...(((.(----------((..((((..((((......)))).))))(((((((((....))))))(((......(((((((.....)))))))...)))..)))...).))))) ( -33.10, z-score = -2.95, R) >droSim1.chr3L 1753828 105 + 22553184 -ACCCAGUG----------CCAAGUGCCCAGGAUGUUAUUCCUAGCGCCGCGCAAACAUCUGUUUGCCACCAACCUGAUCAUCCUGCUGAUGAUCUACGUGAUGCGCAAGUGCCUG -...(((.(----------((..((((..((((......)))).))))(((((((((....))))))(((......(((((((.....)))))))...)))..)))...).))))) ( -33.10, z-score = -2.87, R) >dp4.chrXR_group6 12110002 108 + 13314419 -ACCCAGUGUG-------CCCAAGUGCUUAGGAUGCUGUUCCUUGCGCCGCGCAAGCAUCUGUUUGCCACCAACCUGAUCAUCCUGCUGAUGAUCUACGUGAUGCGCAAGUGCCUG -...(((.(..-------(....((((..((((......)))).)))).(((((.(((......)))(((......(((((((.....)))))))...))).)))))..)..)))) ( -35.80, z-score = -2.50, R) >droWil1.scaffold_180916 1515217 115 - 2700594 -ACCCAGUGCUUCCCCAUCCUCAAGGCUCAGGAUGUUAUUCCUAGCGCCGCGAAAGCAUCUCUUUGCCACUAAUUUGAUCAUGCUUCUUGUGAUCUACAUCAUGCGCAAGUGCUUG -....((..(((...((((((........)))))).........((((.((....))...................((((((.......))))))........)))))))..)).. ( -29.30, z-score = -1.59, R) >droMoj3.scaffold_6680 8907928 107 + 24764193 UGCCCAGUGU---------CUAACCAGUCAGGAUGUUGUUUCUGGCGCCGCGCAAGCAUCUAUUUGCCACCAAUUUGAUAAUAUUCCUCAUGAUUUAUAUCAUGCGCAAGUGCCUG ....(((..(---------((.........)))..))).....(((((.(((((.(((......)))........(((((....((.....))....))))))))))..))))).. ( -25.10, z-score = -0.49, R) >droVir3.scaffold_13049 13317332 107 - 25233164 UGCCCAGUGU---------AUAAUCAGCCAGGAUGUUAUUUCUGGCGCCGCGCAAGCAUCUAUUUGCCAGCAAUUUGAUAAUAUUUCUAUUGAUUUAUAUCAUGCGCAAGUGCCUG ....(((.((---------((.....(((((((......)))))))...(((((.(((......)))........(((((.((..((....))..))))))))))))..))))))) ( -31.60, z-score = -2.10, R) >droGri2.scaffold_15110 13204660 107 - 24565398 UGCCCAGUGU---------AUUCCCGGUCAGAAUGUUAUUUCUGGCGCCGCGCAAGCAUCUACUGGCGAGCAAUUUGAUAAUUUUGCUAAUGAUUUAUAUCAUGCGCAAGUGUCUC (((((((((.---------.......(((((((......)))))))...((....))...))))))...)))....((((...((((..(((((....)))))..)))).)))).. ( -32.10, z-score = -1.83, R) >consensus _ACCCAGUG__________CCAAGUGCCCAGGAUGUUAUUCCUAGCGCCGCGCAAACAUCUGUUUGCCACCAACCUGAUCAUCCUGCUGAUGAUCUACGUGAUGCGCAAGUGCCUG .............................((((......)))).((((((((((((......))))).........(((.(((.....))).)))........)))...))))... (-14.56 = -13.78 + -0.78)
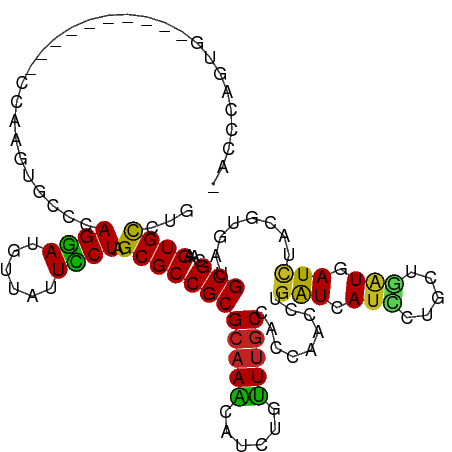
| Location | 2,197,543 – 2,197,657 |
|---|---|
| Length | 114 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.69 |
| Shannon entropy | 0.40686 |
| G+C content | 0.50333 |
| Mean single sequence MFE | -35.88 |
| Consensus MFE | -14.42 |
| Energy contribution | -14.05 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.43 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.686563 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 2197543 114 + 24543557 UAUUCCUAGCGCCGCGCAAACAUCUGUUUGCCACCAACCUGAUCAUCCUGCUGAUGAUCUACUUGAUGCGCAAGUGCCUGCAGCUCUUCUGCAUCAUCGAGAACCGCGCCGUUC------ .......((((.(((((((((....)))))).........(((((((.....)))))))..(((((((.((....)).(((((.....))))).)))))))....))).)))).------ ( -35.80, z-score = -3.20, R) >droAna3.scaffold_13337 22653751 114 - 23293914 UGUUCCUGGCGCCGCGCAAACACCUGUUUGCCACCAACCUGGUUAUCCUGCUGAUGAUCUAUGUGAUGCGCAAGUGCCUCCAGCUGUUCUGCAUAAUCGAGAACGGCACCACCG------ .......((((((((((((((....))))))(((......(((((((.....)))))))...)))..)))...)))))....((((((((.(......))))))))).......------ ( -35.80, z-score = -1.97, R) >droEre2.scaffold_4784 2192418 114 + 25762168 UAUUCCUCGCGCCGCGCAAACAUCUGUUUGCCACCAACUUGAUCAUCCUGCUGAUGAUCUACGUGAUGCGCAAGUGCCUGCAGCUCUUCUGCAUCAUCGAGAACCGCACCGCUC------ ........(((..((((((((....))))))......((((((((((.....)))))))...((((((((.(((.((.....)).))).)))))))).)))....))..)))..------ ( -36.40, z-score = -3.15, R) >droYak2.chr3L 2150965 114 + 24197627 UAUUCCUAGCGCCGCGCAAACAUCUGUUUGCCACCAACCUAAUCAUCCUUGUGGUGAUCUACGUGAUGCGAAAGUGCCUGCAGCUCUUCUGCAUCAUCGAGAACCGCGCAGUUC------ ........((((...((((((....))))))(((((...............))))).(((.(((((((((.(((.((.....)).))).))))))).)))))...)))).....------ ( -35.86, z-score = -2.90, R) >droSec1.super_2 2223560 114 + 7591821 UAUUCCUAGCGCCGCGCAAACAUCUGUUUGCCACUAACCUGAUCAUCCUGCUGAUGAUCUACGUGAUGCGCAAGUGCCUGCAGCUCUUCUGCAUCAUCGAGAACCGCGCCGUUC------ .......((((.(((((((((....))))))..((.....(((((((.....)))))))..(((((((((.(((.((.....)).))).))))))).))))....))).)))).------ ( -37.80, z-score = -3.40, R) >droSim1.chr3L 1753853 114 + 22553184 UAUUCCUAGCGCCGCGCAAACAUCUGUUUGCCACCAACCUGAUCAUCCUGCUGAUGAUCUACGUGAUGCGCAAGUGCCUGCAGCUCUUCUGCAUCAUCGAGAACCGCGCCGUUC------ .......((((.(((((((((....)))))).........(((((((.....)))))))..(((((((((.(((.((.....)).))).))))))).))......))).)))).------ ( -37.70, z-score = -3.36, R) >dp4.chrXR_group6 12110030 114 + 13314419 UGUUCCUUGCGCCGCGCAAGCAUCUGUUUGCCACCAACCUGAUCAUCCUGCUGAUGAUCUACGUGAUGCGCAAGUGCCUGCAGCUCUUCUGCAUCCUCGAGAACCGCACCGCCU------ .((((((((((((((((((((....)))))).........(((((((.....)))))))...)))..)))))))....(((((.....))))).......))))..........------ ( -35.90, z-score = -2.45, R) >droWil1.scaffold_180916 1515252 120 - 2700594 UAUUCCUAGCGCCGCGAAAGCAUCUCUUUGCCACUAAUUUGAUCAUGCUUCUUGUGAUCUACAUCAUGCGCAAGUGCUUGCAGCUAUUUUGUAUUAUUGAGAACAAGACUGGAGCCGAUG ........((.((((....)).((((..............((((((.......))))))((((....((((((....)))).)).....)))).....))))........)).))..... ( -27.00, z-score = 0.29, R) >droMoj3.scaffold_6680 8907955 118 + 24764193 UGUUUCUGGCGCCGCGCAAGCAUCUAUUUGCCACCAAUUUGAUAAUAUUCCUCAUGAUUUAUAUCAUGCGCAAGUGCCUGCAGUUGUUCUGCAUCAUCGAGAACAAGGCGUGCCAGCA-- .....(((((((.((....))....((((((.........((......))..((((((....)))))).))))))((((((((.....)))).((.....))...)))))))))))..-- ( -35.60, z-score = -1.38, R) >droVir3.scaffold_13049 13317359 118 - 25233164 UAUUUCUGGCGCCGCGCAAGCAUCUAUUUGCCAGCAAUUUGAUAAUAUUUCUAUUGAUUUAUAUCAUGCGCAAGUGCCUGCAGUUGUUCUGCAUCAUUGAGAACAAGGCAUGCCAGAA-- ...(((((((...(((((.(((......)))........(((((.((..((....))..))))))))))))..((((((.....((((((.((....)))))))))))))))))))))-- ( -40.20, z-score = -3.06, R) >droGri2.scaffold_15110 13204687 118 - 24565398 UAUUUCUGGCGCCGCGCAAGCAUCUACUGGCGAGCAAUUUGAUAAUUUUGCUAAUGAUUUAUAUCAUGCGCAAGUGUCUCCAAUUGUUCUGCAUCAUUGAGAACAAGGCGUGCAAGAA-- ...((((.(((((((....))...((((.((((((((..(....)..))))).(((((....))))).))).)))).......(((((((.((....))))))))))))))...))))-- ( -36.60, z-score = -1.70, R) >consensus UAUUCCUAGCGCCGCGCAAACAUCUGUUUGCCACCAACCUGAUCAUCCUGCUGAUGAUCUACGUGAUGCGCAAGUGCCUGCAGCUCUUCUGCAUCAUCGAGAACCGCGCCGGCC______ ........((((((((((((......))))).........(((.(((.....))).)))........)))...)))).(((((.....)))))........................... (-14.42 = -14.05 + -0.37)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:57:15 2011