| Sequence ID | dm3.chr3L |
|---|---|
| Location | 2,196,475 – 2,196,569 |
| Length | 94 |
| Max. P | 0.625173 |

| Location | 2,196,475 – 2,196,569 |
|---|---|
| Length | 94 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 69.17 |
| Shannon entropy | 0.60160 |
| G+C content | 0.34417 |
| Mean single sequence MFE | -18.39 |
| Consensus MFE | -8.68 |
| Energy contribution | -9.41 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.625173 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
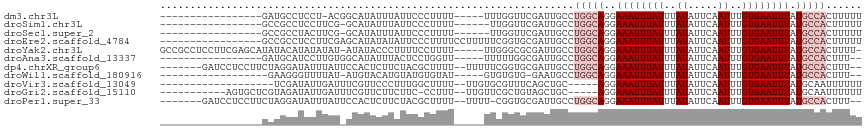
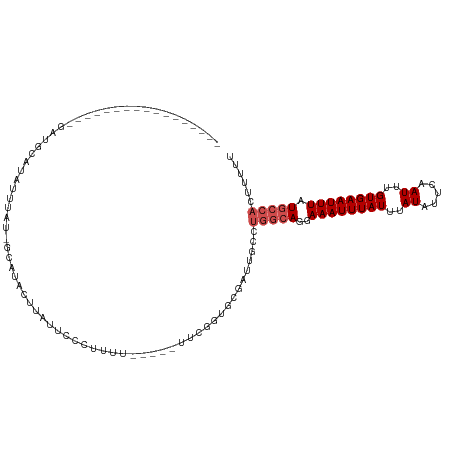
>dm3.chr3L 2196475 94 + 24543557 -----------------GAUGCCUCCU-ACGGCAUAUUUAUUCCCUUUU-----UUUGGUUCGAUUGCCUGGCAGGAAAUUUAUUUAUAUUCAAUUUGUGAAUUUAUGCCACUUUUU -----------------.(((((....-..)))))..............-----...(((......)))(((((..((((((((..((.....))..)))))))).)))))...... ( -20.40, z-score = -2.23, R) >droSim1.chr3L 1752826 93 + 22553184 -----------------GCCGCCUCCUUCG-GCAUAUUUAUUCCCUUUU------UUGGUUCGAUUGCCUGGCAGGAAAUUUAUUUAUAUUCAAUUUGUGAAUUUAUGCCACUUUUU -----------------((((.......))-))................------..(((......)))(((((..((((((((..((.....))..)))))))).)))))...... ( -19.30, z-score = -1.61, R) >droSec1.super_2 2222515 93 + 7591821 -----------------GCCGCCUACUUCG-GCAUAUUUAUUCCCUUUU------UUGGUUCGAUUGCCUGGCAGGAAAUUUAUUUAUAUUCAAUUUGUGAAUUUAUGCCACUUUUU -----------------((((.......))-))................------..(((......)))(((((..((((((((..((.....))..)))))))).)))))...... ( -19.30, z-score = -1.79, R) >droEre2.scaffold_4784 2191191 100 + 25762168 -----------------GCCGCCUCCUUCGAGCAUAUAUAUUCCCUUUUCCUUUUUCGGUGCGAUUGCCUGGCAGGAAAUUUAUUUAUAUUCAAUUUGUGAAUUUAUGCCACUUUUU -----------------............(.(((((..........((((((...((((.((....)))))).))))))...(((((((.......))))))).))))))....... ( -16.00, z-score = -0.12, R) >droYak2.chr3L 2149844 110 + 24197627 GCCGCCUCCUUCGAGCAUAUACAUAUAUAU-AUAUACCCUUUUCCUUUU-----UUGGGCGCGAUUGCCUGGCAGGAAAUUUAUUUAUAUUCAAUUUGUGAAUUUAUGCCACUUUU- ............(.(((((((((.....((-(((......((((((...-----(..((((....))))..).))))))......)))))......))))....))))))......- ( -20.80, z-score = -0.89, R) >droAna3.scaffold_13337 22652931 93 - 23293914 -----------------GAUGCAUCCUUGUGGCAUAUUUACUCCUGGUU-----UUUUUGGCGAUUGCCUGGCAGGAAAUUUAUUUAUAUUCAAUUUGUGAAUUUAUGCCACUUU-- -----------------...........((((((((.....(((((.(.-----.....(((....))).).))))).....(((((((.......))))))).))))))))...-- ( -23.60, z-score = -2.17, R) >dp4.chrXR_group6 12108799 106 + 13314419 -------GAUCCUCCUUCUAGGAUAUUUAUUCCACUCUUCUACGCUUUU--UUUUUCGGUGCGAUUGCCUGGCAGGAAAUUUAUUUAUAUUCAAUUUGUGAAUUUAUGCCACUUU-- -------.(((((......)))))..................(((....--.........)))......(((((..((((((((..((.....))..)))))))).)))))....-- ( -18.22, z-score = -1.04, R) >droWil1.scaffold_180916 1513565 90 - 2700594 ------------------GAAGGGUUUUAU-AUGUACAUGUAUGUGUAU-----GUGUGUG-GAAUGCCUGGCAGGAAAUUUAUUUAUAUUCAAUUUGUGAAUUUAUGCCACUUU-- ------------------..((((((((((-((((((((....))))))-----..)))))-)))).)))((((..((((((((..((.....))..)))))))).)))).....-- ( -23.20, z-score = -2.18, R) >droVir3.scaffold_13049 13316744 91 - 25233164 -------------------UCGAUAUUGAUUUCGUUCCCUUUGGCUUUU--UUGUGCGUUUCAGCUGC-----AGGAAAUUUAUUUAUAUUCAAUUUGUGAAUUUAUGCAAUUUUUU -------------------.(((........))).........((.(((--((((((......)).))-----)))))((..(((((((.......)))))))..))))........ ( -11.30, z-score = 0.95, R) >droGri2.scaffold_15110 13204106 98 - 24565398 -----------AGUGCUCGUAGAUAUUGAUUUCGUUCUUCUUC-CCUUU--UUGUUCGCUGUAGCUGC-----AGGAAAUUUAUUUAUAUUCAAUUUGUGAAUUUAUGCAAUUUUUU -----------..(((..(((((((..(((((((..(......-.....--..)..).((((....))-----)))))))))))))))(((((.....)))))....)))....... ( -11.82, z-score = 0.78, R) >droPer1.super_33 132007 105 + 967471 -------GAUCCUCCUUCUAGGAUAUUUAUUCCACUCUUCUACGCUUUU--UUUU-CGGUGCGAUUGCCUGGCAGGAAAUUUAUUUAUAUUCAAUUUGUGAAUUUAUGCCACUUU-- -------.(((((......)))))..................(((....--....-....)))......(((((..((((((((..((.....))..)))))))).)))))....-- ( -18.30, z-score = -1.11, R) >consensus _________________GAUGCAUAUUUAU_GCAUACUUAUUCCCUUUU_____UUCGGUGCGAUUGCCUGGCAGGAAAUUUAUUUAUAUUCAAUUUGUGAAUUUAUGCCACUUUUU .....................................................................(((((..((((((((..((.....))..)))))))).)))))...... ( -8.68 = -9.41 + 0.73)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:57:13 2011