| Sequence ID | dm3.chr3L |
|---|---|
| Location | 2,159,233 – 2,159,351 |
| Length | 118 |
| Max. P | 0.904482 |

| Location | 2,159,233 – 2,159,351 |
|---|---|
| Length | 118 |
| Sequences | 9 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.37 |
| Shannon entropy | 0.57062 |
| G+C content | 0.47601 |
| Mean single sequence MFE | -33.89 |
| Consensus MFE | -12.70 |
| Energy contribution | -13.09 |
| Covariance contribution | 0.38 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.17 |
| SVM RNA-class probability | 0.904482 |
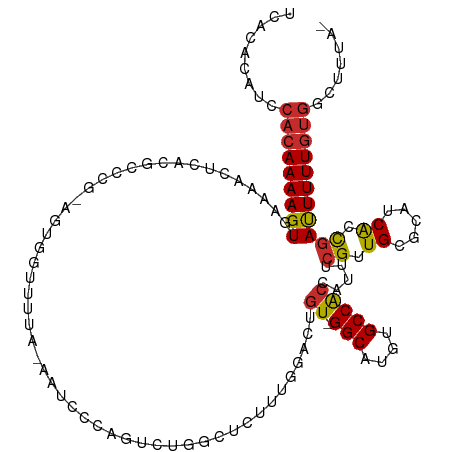
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 2159233 118 + 24543557 UCACACAUCCACAAAAGUGAAAACUCACGCCCGGAGUGGUUUUUUAAUCCCAGUCUAGCUCUUUGGACUGU-GGCAUGUGCCACCAUUUCGAUGCGCAUCACCGAUUUUUGUGGCUGUA- ....(((.((((((((((((....))))((.(((((((((..........((((((((....)))))))).-(((....))))))))))))..))...........)))))))).))).- ( -41.40, z-score = -3.77, R) >droSim1.chr3L 1715032 117 + 22553184 UCACACAUCCACAAAAGUGAAAACUCACGCCCGGAGUGGUUUUA-AAUCGCCGUCUGGCUCUUUGGACUGU-GGCAUGUGCCACCAUUUCGUUGCGCAUCACCGAUUUUUGUGGCUGUA- ....(((.((((((((((((....))))((.(((((((((....-....((.(((..(....)..))).))-(((....))))))))))))..))...........)))))))).))).- ( -38.70, z-score = -2.10, R) >droSec1.super_2 2185295 117 + 7591821 UCACACAUCCACAAAAGUGAAAACUCACGCCCGAAGUGGUUUUA-AAUCGCAGUCUGGCUCUUUGGACUGU-GGCAUGUGCCACCAUUUCGUUGCGCAUCACCGAUUUUUGUGGCUGUA- ....(((.((((((((((((....))))((.(((((((((....-....((((((..(....)..))))))-(((....))))))))))))..))...........)))))))).))).- ( -43.80, z-score = -4.02, R) >droYak2.chr3L 2112255 117 + 24197627 UCACACAUCCACAAAAGUGAAAACUCACGCCCGGAGUGGUUUUA-AAUCCCAGUCUGGCUCUUUGGACUGU-GGCAUGUGCCACCAUUUCGUUGCGCAUCACCGAUUUUUGUGGCUUUA- ........((((((((((((....))))((.(((((((((....-.....(((((..(....)..))))).-(((....))))))))))))..))...........)))))))).....- ( -38.00, z-score = -2.61, R) >droEre2.scaffold_4784 2154222 117 + 25762168 UCACACAUCCACAAAAGUGAAAACUCACGCCCGGAGCGGUUUUU-AAUCCCAGUCUGACUCUUUGGACUGU-GGCAUGUGCCACCAUUUCGCUGCGCAUCACCAAUUUUUGUGGCUUUA- ........((((((((((((....))))...((.(((((.....-......((((..(....)..))))((-(((....)))))....))))).))..........)))))))).....- ( -34.80, z-score = -2.42, R) >droAna3.scaffold_13337 22619797 113 - 23293914 UCACACAUUCACAAAAGUGAAACCUCAUCGG-----AGGUCUACGACUCCCUGUCUGGCUCUGGUGCUUGU-GGCAUGUGCCACCAUUUCGUUGUGCAUCGCCGACUUUUGUGGCUUGA- .....((.((((((((((...(((((....)-----))))....(((.....)))((((...(((((..((-(((....)))))((......)).)))))))))))))))))))..)).- ( -36.60, z-score = -1.58, R) >dp4.chrXR_group6 12068903 90 + 13314419 ----UCUCACACAAAAGUGAAACCUC---------------------UCAUUCUGUGGAUUGGAU---UGU-GGCAUGUGCCGCCAUUUCGUUGCGCAUCAACGACUUUUGUGGCUUUA- ----.....(((((((((((((((..---------------------((........))..))..---.((-(((....)))))..))))((((.....)))).)))))))))......- ( -24.70, z-score = -1.22, R) >droPer1.super_33 91269 90 + 967471 ----UCUCACACAAAAGUGAAACCUC---------------------UCAUUCUGUGGAUUGGAU---UGU-GGCAUGUGCCACCAUUUCGUUGCGCAUCAACGACUUUUGUGGCUUUA- ----.....(((((((((((((((..---------------------((........))..))..---.((-(((....)))))..))))((((.....)))).)))))))))......- ( -25.10, z-score = -1.58, R) >droWil1.scaffold_180916 1469373 99 - 2700594 ------------AAAAGUGAAAUCCCUUCC------UGGUCUAGAAGUCAC--UCUGUCUAGUCGGAGCGUUGGCAUGUGCCAGU-UUUCGUUGCGCAUCAAUGAUUUUUAUAGCCGCCA ------------...((.(((.....))))------)(((.(((((((((.--((((......))))((((((((....))))..-.......)))).....)))))))))..))).... ( -21.90, z-score = -0.01, R) >consensus UCACACAUCCACAAAAGUGAAAACUCACGCCCG_AGUGGUUUUA_AAUCCCAGUCUGGCUCUUUGGACUGU_GGCAUGUGCCACCAUUUCGUUGCGCAUCACCGAUUUUUGUGGCUUUA_ .........(((((((((((....))...............................................(.((((((.((......)).)))))))....)))))))))....... (-12.70 = -13.09 + 0.38)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:57:10 2011