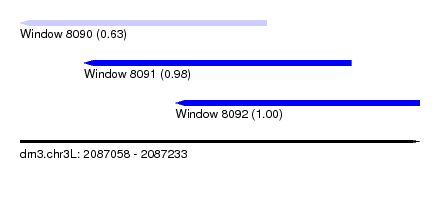
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 2,087,058 – 2,087,233 |
| Length | 175 |
| Max. P | 0.999596 |

| Location | 2,087,058 – 2,087,166 |
|---|---|
| Length | 108 |
| Sequences | 11 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 73.33 |
| Shannon entropy | 0.57435 |
| G+C content | 0.57967 |
| Mean single sequence MFE | -37.98 |
| Consensus MFE | -15.21 |
| Energy contribution | -14.42 |
| Covariance contribution | -0.79 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.632320 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
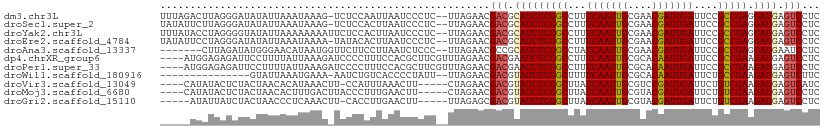
>dm3.chr3L 2087058 108 - 24543557 UUCAAUUGCGAACGAUUGAUUCCGCCGAGGAUGAGUCCUCUUCCUCUGGCCGGGAGGACAGUGCCCCCGAGUCGCCACACGCUUUCCAACCUGGCCAGCAACCAGGCC--- .(((((((....)))))))....((((((((....))))).....(((((((((.(((.((((................)))).)))..)))))))))......))).--- ( -39.89, z-score = -1.95, R) >droSec1.super_2 2117978 108 - 7591821 UUCAAUUGCGAACGAUUGAUUCCGCCGAGGAUGAGUCCUCUUCCUCUGGCCGGGAGGACAGUGCCCCCGAGUCGCCACACGCUUUCCAACCUGGCCAGCAACCAGGCC--- .(((((((....)))))))....((((((((....))))).....(((((((((.(((.((((................)))).)))..)))))))))......))).--- ( -39.89, z-score = -1.95, R) >droYak2.chr3L 2044860 108 - 24197627 UUCAAUUGCGAACGAUUGAUUCCGCCGAGGAUGAGUCCUCUUCCUCUGGCCGGGAGGACAGUGCCCCCGAGUCGCCACACGCUUUCCAACCUGGCCAGCAACCCGGCC--- .(((((((....)))))))....((((((((....))))).....(((((((((.(((.((((................)))).)))..)))))))))......))).--- ( -39.99, z-score = -1.96, R) >droEre2.scaffold_4784 2085930 108 - 25762168 UUCAAUUGCGAACGAUUGAUUCCGCCGAGGAUGAGUCCUCUUCCUCUGGCCGGGAGGACAGUGCCCCCGAGUCGCCACACGCUUUCCAACCUGGCCAGCAACCCGGCC--- .(((((((....)))))))....((((((((....))))).....(((((((((.(((.((((................)))).)))..)))))))))......))).--- ( -39.99, z-score = -1.96, R) >droAna3.scaffold_13337 22555368 111 + 23293914 AUCAAUUGCGAACGAUUGAUUCCGCCGAGGAUGAAUCCUCGUCUUCUGGUCGGGAGGAGAGCGGUCCGGAGUCGCCGAGUGCUUUCCAUCCUGGCCAGCAACCUGGAGGUC .(((((((....)))))))(((((.((((((....)))))).)..((((((((((((((((((.((.((.....)))).)))))))).))))))))))......))))... ( -51.90, z-score = -3.16, R) >dp4.chrXR_group6 11996233 108 - 13314419 UUCAAUUGCGCACAAUUGAUUCCGCCGAAGAUGAGUCCUCAUCCUCCGGCCGGGAGGACG---CACCCGACUCGCCCCUGCAGCCAGGACAACAGCUGCAGCCGGGGGGCC .(((((((....)))))))....(((...(((((....)))))(((((((((((......---..)))).........((((((..(.....).)))))))))))))))). ( -42.90, z-score = -1.32, R) >droPer1.super_33 17186 108 - 967471 UUCAAUUGCGCACAAUUGAUUCCGCCGAAGAUGAGUCCUCAUCCUCCGGCCGGGAGGAUG---CACCCGACUCGCCCCUGCAGCCAGGACAACAGCUGCAGCCGGGAGGCC .(((((((....)))))))....(((......(((((..((((((((.....))))))))---.....))))).((((((((((..(.....).)))))))..))).))). ( -45.20, z-score = -2.62, R) >droWil1.scaffold_180916 1392905 111 + 2700594 UUCAAUUGCGCACAAUUGAUUCUGCCGAAGAUGAGUCUUCUUCUUCUGGCCGGGAAGAUAGUGCACCUGAAUCACAAUAUCCCCAGCAGCCAGGUCAACAACAGCCCGGCC .(((((((....))))))).......(((((.(......).))))).(((((((.......((.(((((.....................))))))).......))))))) ( -32.44, z-score = -1.53, R) >droVir3.scaffold_13049 13211313 111 + 25233164 AUCAAUUGCGUCCGAUUGAUUCUGUCGAAGAUGAGUCAUCAUCGUCUGGCAGGGAAGAUAGCUCUCCUGAAUCGCCGCAUCAGCAACAGCAGUCACAACAGCAGACAGGCG ((((((((....))))))))((((((...(((((....)))))(.(((.((((..((....))..)))).......((....)).....))).).........)))))).. ( -31.00, z-score = -0.11, R) >droMoj3.scaffold_6680 8798731 108 - 24764193 AUCAAUUGCGUACGAUUGAUUCUGUCGAAGAUGAGUCCUCGUCAUCUGGCAGGGAAGAUAGUUCACCUGAAUCGCCCAACCAGUUCCAGCAG---CAACAGCAGCCGGGCG .(((((((....)))))))(((((((((.(((((....))))).)).)))))))......((((....))))(((((.....((....)).(---(....))....))))) ( -30.30, z-score = -0.41, R) >droGri2.scaffold_15110 13103391 105 + 24565398 AUCAAUUGCGUACGAUUGAUUCUGUCGAAGAUGAGUCCUCGUCAUCUGGCAAGGAUGAUAGCUCGCCUGAAUCACCCCAAAAUUACCAGC---AGCAGCAACCUGGCC--- ((((((((....))))))))......((.(((((....))))).)).(((.(((.((...(((.((.((.................))))---)))..)).))).)))--- ( -24.23, z-score = 0.10, R) >consensus UUCAAUUGCGAACGAUUGAUUCCGCCGAAGAUGAGUCCUCUUCCUCUGGCCGGGAGGACAGUGCACCCGAGUCGCCACACCCUUUCCAACCUGGCCAGCAACCGGGCGG__ .(((((((....)))))))(((((((((.((.((....)).)).)).))).))))........................................................ (-15.21 = -14.42 + -0.79)

| Location | 2,087,086 – 2,087,203 |
|---|---|
| Length | 117 |
| Sequences | 11 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 77.61 |
| Shannon entropy | 0.48418 |
| G+C content | 0.53133 |
| Mean single sequence MFE | -35.64 |
| Consensus MFE | -27.39 |
| Energy contribution | -26.94 |
| Covariance contribution | -0.45 |
| Combinations/Pair | 1.26 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.77 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.976844 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 2087086 117 - 24543557 --UCCAAUUAAUCCCUCUUAGAACGACGCAUCUCGGCCUUUCAAUUGCGAACGAUUGAUUCCGCCGAGGAUGAGUCCUCUUCCUCUGGCCGGGAGGACAGUGCCCCCGAGUCGCCACAC --.................(((..(((.(((((((((...(((((((....)))))))....))))).)))).))).))).....((((((((.((......))))))....))))... ( -36.40, z-score = -1.24, R) >droSec1.super_2 2118006 117 - 7591821 --UCCACUUAAUCCCUCUUAGAACGACGCAUCUCGGCCUUUCAAUUGCGAACGAUUGAUUCCGCCGAGGAUGAGUCCUCUUCCUCUGGCCGGGAGGACAGUGCCCCCGAGUCGCCACAC --.................(((..(((.(((((((((...(((((((....)))))))....))))).)))).))).))).....((((((((.((......))))))....))))... ( -36.40, z-score = -1.19, R) >droYak2.chr3L 2044888 117 - 24197627 --UCCACUUAAUCCCUCUUAGAACGACGCAUCUCGGCCUUUCAAUUGCGAACGAUUGAUUCCGCCGAGGAUGAGUCCUCUUCCUCUGGCCGGGAGGACAGUGCCCCCGAGUCGCCACAC --.................(((..(((.(((((((((...(((((((....)))))))....))))).)))).))).))).....((((((((.((......))))))....))))... ( -36.40, z-score = -1.19, R) >droEre2.scaffold_4784 2085958 117 - 25762168 --UACACUUAAUCCCUCUUAGAACGACGCAUCUCGGCCUUUCAAUUGCGAACGAUUGAUUCCGCCGAGGAUGAGUCCUCUUCCUCUGGCCGGGAGGACAGUGCCCCCGAGUCGCCACAC --.................(((..(((.(((((((((...(((((((....)))))))....))))).)))).))).))).....((((((((.((......))))))....))))... ( -36.40, z-score = -1.45, R) >droAna3.scaffold_13337 22555399 117 + 23293914 --CUUCCUUAAUCUCCCUUAGAACGCCGCAUCUCGGCCUAUCAAUUGCGAACGAUUGAUUCCGCCGAGGAUGAAUCCUCGUCUUCUGGUCGGGAGGAGAGCGGUCCGGAGUCGCCGAGU --.((((....(((.....)))..(((((.((((......(((((((....)))))))(((((((((((((((....)))))))).)).))))).)))))))))..))))......... ( -40.00, z-score = -0.10, R) >dp4.chrXR_group6 11996266 114 - 13314419 CCCUUUCCACGCUUCGUUUAGAACGACGAAUCUCGGCCUUUCAAUUGCGCACAAUUGAUUCCGCCGAAGAUGAGUCCUCAUCCUCCGGCCGGGAGGA---CGCACCCGACUCGCCCC-- ..........((.(((((......)))))((((((((...(((((((....)))))))....)))).))))(((((..(.((((((.....))))))---.).....)))))))...-- ( -36.10, z-score = -2.04, R) >droPer1.super_33 17219 114 - 967471 CCCUUUCCACGCUUCGUUUAGAACGACGAAUCUCGGCCUUUCAAUUGCGCACAAUUGAUUCCGCCGAAGAUGAGUCCUCAUCCUCCGGCCGGGAGGA---UGCACCCGACUCGCCCC-- ..........((.(((((......)))))((((((((...(((((((....)))))))....)))).))))(((((..((((((((.....))))))---)).....)))))))...-- ( -40.40, z-score = -3.41, R) >droWil1.scaffold_180916 1392936 117 + 2700594 --UCUGUCACCCCUAUUUUAGAACGACGUAUCUCGGCUUUUCAAUUGCGCACAAUUGAUUCUGCCGAAGAUGAGUCUUCUUCUUCUGGCCGGGAAGAUAGUGCACCUGAAUCACAAUAU --.(((((.((((((....((((.(((.(((((((((...(((((((....)))))))....)))).))))).))))))).....)))..)))..)))))................... ( -33.10, z-score = -2.43, R) >droVir3.scaffold_13049 13211344 115 + 25233164 ----UCCAUUUAAACUUCUAGAACGACGUAUCUCGGCUUAUCAAUUGCGUCCGAUUGAUUCUGUCGAAGAUGAGUCAUCAUCGUCUGGCAGGGAAGAUAGCUCUCCUGAAUCGCCGCAU ----.............(((((..(((.(((((((((..((((((((....))))))))...)))).))))).))).......)))))((((..((....))..))))........... ( -30.20, z-score = -0.36, R) >droMoj3.scaffold_6680 8798759 116 - 24764193 ---UACCCUUUGAACUUCUAGAACGACGUAUCUCGGCUUAUCAAUUGCGUACGAUUGAUUCUGUCGAAGAUGAGUCCUCGUCAUCUGGCAGGGAAGAUAGUUCACCUGAAUCGCCCAAC ---.......(((((((((.....(((.(((((((((..((((((((....))))))))...)))).))))).)))((((((....))).))).))).))))))............... ( -31.60, z-score = -1.24, R) >droGri2.scaffold_15110 13103416 115 + 24565398 ----UCACCUUGAACUUUUAGAGCGACGUAUCUCGGCUUAUCAAUUGCGUACGAUUGAUUCUGUCGAAGAUGAGUCCUCGUCAUCUGGCAAGGAUGAUAGCUCGCCUGAAUCACCCCAA ----......(((.......(((((((.(((((((((..((((((((....))))))))...)))).))))).)))...(((((((.....))))))).)))).......)))...... ( -35.04, z-score = -2.26, R) >consensus __UCUACCUAAUCCCUUUUAGAACGACGCAUCUCGGCCUUUCAAUUGCGAACGAUUGAUUCCGCCGAAGAUGAGUCCUCUUCCUCUGGCCGGGAGGACAGUGCACCCGAGUCGCCACAC ....................((..(((.(((((((((...(((((((....)))))))....))))).)))).)))....((((((.....)))))).............))....... (-27.39 = -26.94 + -0.45)


| Location | 2,087,126 – 2,087,233 |
|---|---|
| Length | 107 |
| Sequences | 11 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 69.98 |
| Shannon entropy | 0.64611 |
| G+C content | 0.42011 |
| Mean single sequence MFE | -26.67 |
| Consensus MFE | -20.86 |
| Energy contribution | -20.66 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.78 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.06 |
| SVM RNA-class probability | 0.999596 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 2087126 107 - 24543557 UUUAGACUUAGGGAUAUAUUAAAUAAAG-UCUCCAAUUAAUCCCUC--UUAGAACGACGCAUCUCGGCCUUUCAAUUGCGAACGAUUGAUUCCGCCGAGGAUGAGUCCUC ......((.((((((.............-..........)))))).--..))...(((.(((((((((...(((((((....)))))))....))))).)))).)))... ( -28.80, z-score = -2.16, R) >droSec1.super_2 2118046 107 - 7591821 UAUAUUCUUAGGGAUAUAUUAAAUAAAG-UCUCCACUUAAUCCCUC--UUAGAACGACGCAUCUCGGCCUUUCAAUUGCGAACGAUUGAUUCCGCCGAGGAUGAGUCCUC ....((((.((((((.............-..........)))))).--..)))).(((.(((((((((...(((((((....)))))))....))))).)))).)))... ( -30.80, z-score = -3.01, R) >droYak2.chr3L 2044928 108 - 24197627 UUUAUACCUAGGGGUAUAUUAAAAAAAAUUCUCCACUUAAUCCCUC--UUAGAACGACGCAUCUCGGCCUUUCAAUUGCGAACGAUUGAUUCCGCCGAGGAUGAGUCCUC .......(((((((...(((((..............)))))..)))--))))...(((.(((((((((...(((((((....)))))))....))))).)))).)))... ( -29.14, z-score = -2.60, R) >droEre2.scaffold_4784 2085998 107 - 25762168 UAUAUUCCUAGGGAUAUAUUAAAUAAAA-UAUACACUUAAUCCCUC--UUAGAACGACGCAUCUCGGCCUUUCAAUUGCGAACGAUUGAUUCCGCCGAGGAUGAGUCCUC ....(((..(((((((((((......))-)))).......))))).--...))).(((.(((((((((...(((((((....)))))))....))))).)))).)))... ( -29.41, z-score = -3.24, R) >droAna3.scaffold_13337 22555439 101 + 23293914 -------CUUAGAUAUGGGAACAUAAUGGUUCUUCCUUAAUCUCCC--UUAGAACGCCGCAUCUCGGCCUAUCAAUUGCGAACGAUUGAUUCCGCCGAGGAUGAAUCCUC -------....(((..((((...(((.((.....)))))...))))--...........(((((((((..((((((((....))))))))...))))).)))).)))... ( -27.10, z-score = -1.56, R) >dp4.chrXR_group6 11996301 106 - 13314419 ----AUGGAGAGAUUCCUUUUAUUAAAGAUCCCCUUUCCACGCUUCGUUUAGAACGACGAAUCUCGGCCUUUCAAUUGCGCACAAUUGAUUCCGCCGAAGAUGAGUCCUC ----.(((((((....((((....)))).....))))))).(.(((.....))))(((..((((((((...(((((((....)))))))....)))).))))..)))... ( -29.90, z-score = -2.85, R) >droPer1.super_33 17254 106 - 967471 ----AUGGAGAGAUUCCUUUUAUUAAAGAUCCCCUUUCCACGCUUCGUUUAGAACGACGAAUCUCGGCCUUUCAAUUGCGCACAAUUGAUUCCGCCGAAGAUGAGUCCUC ----.(((((((....((((....)))).....))))))).(.(((.....))))(((..((((((((...(((((((....)))))))....)))).))))..)))... ( -29.90, z-score = -2.85, R) >droWil1.scaffold_180916 1392976 92 + 2700594 ---------------GUAUUAAAUGAAA-AAUCUGUCACCCCUAUU--UUAGAACGACGUAUCUCGGCUUUUCAAUUGCGCACAAUUGAUUCUGCCGAAGAUGAGUCUUC ---------------.............-..((((...........--.))))..(((.(((((((((...(((((((....)))))))....)))).))))).)))... ( -22.80, z-score = -2.95, R) >droVir3.scaffold_13049 13211384 100 + 25233164 ----CAUAUACUCUACUAACACAUAAACUU-CCAUUUAAACUU-----CUAGAACGACGUAUCUCGGCUUAUCAAUUGCGUCCGAUUGAUUCUGUCGAAGAUGAGUCAUC ----..........................-............-----.......(((.(((((((((..((((((((....))))))))...)))).))))).)))... ( -20.90, z-score = -2.28, R) >droMoj3.scaffold_6680 8798799 101 - 24764193 ----CAUAUACUCUACUAACACUUUGACUUACCCUUUGAACUU-----CUAGAACGACGUAUCUCGGCUUAUCAAUUGCGUACGAUUGAUUCUGUCGAAGAUGAGUCCUC ----.......................................-----.......(((.(((((((((..((((((((....))))))))...)))).))))).)))... ( -20.40, z-score = -1.68, R) >droGri2.scaffold_15110 13103456 99 + 24565398 -----AUAUUAUCUACUAACCCUCAAACUU-CACCUUGAACUU-----UUAGAGCGACGUAUCUCGGCUUAUCAAUUGCGUACGAUUGAUUCUGUCGAAGAUGAGUCCUC -----.................((((....-....))))....-----...(((.(((.(((((((((..((((((((....))))))))...)))).))))).)))))) ( -24.20, z-score = -2.82, R) >consensus _____ACAUAAGGUUCUAUUAAAUAAAG_UCCCCAUUUAACCCCUC__UUAGAACGACGCAUCUCGGCCUUUCAAUUGCGAACGAUUGAUUCCGCCGAAGAUGAGUCCUC .......................................................(((.(((((((((...(((((((....)))))))....))))).)))).)))... (-20.86 = -20.66 + -0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:57:03 2011