| Sequence ID | dm3.chr3L |
|---|---|
| Location | 2,037,532 – 2,037,652 |
| Length | 120 |
| Max. P | 0.957642 |

| Location | 2,037,532 – 2,037,652 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.78 |
| Shannon entropy | 0.47252 |
| G+C content | 0.42361 |
| Mean single sequence MFE | -30.15 |
| Consensus MFE | -15.10 |
| Energy contribution | -14.55 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.55 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.64 |
| SVM RNA-class probability | 0.957642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
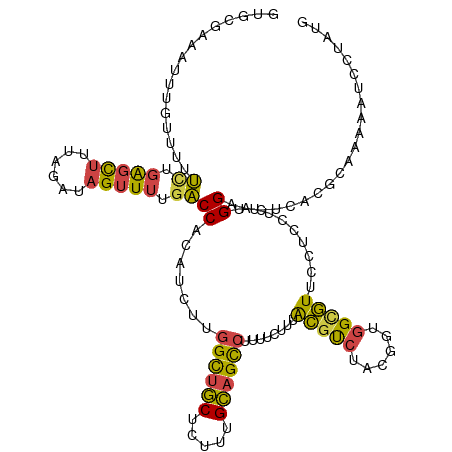
>dm3.chr3L 2037532 120 + 24543557 GUGCGAAAUUUGUUUUUCUGAGCUUUAGAUAGUUUUGACCACAUCUUGGCUGCUCUUUGCAGCCUUUUCUUUACGUCUACGAUGGUGUUCCUCCUUUGGUACUCGCGCAAAAAUCCAAUG ((((((...........(.(((((......))))).)((((......((((((.....)))))).........((....)).))))((.((......)).))))))))............ ( -29.50, z-score = -2.12, R) >droAna3.scaffold_13337 22495837 120 - 23293914 UUACGGUAUCGAUCUUGAUCGUCCUUUGAUAGUUUAACCCAUUCUUUGGUACCUUUUUGGAGUGAAAGCAGCGUCCCCAUAUUUGAAUUAUUUCUUCGGAACUCACGCAUGAAACCAAGG ....(((((((((....)))(((....))).................))))))..(((((.((....)).((((...((....))......(((....)))...))))......))))). ( -21.80, z-score = 0.67, R) >droEre2.scaffold_4784 2032400 120 + 25762168 AUUCGAAAUUUGCUUUUUUGAGCUUCCGUGAGUUUUGACCACAUCUUGGCUGCUCUUUGCAGCCUUUUCUUUACGUCUAAGCUGGCGUUUCUUCUAUGGUACUCGCGCAGAAAUCCUAUG .(((.......((((....))))...(((((((....((((......((((((.....))))))........(((((......)))))........)))))))))))..)))........ ( -33.70, z-score = -3.06, R) >droYak2.chr3L 1994235 120 + 24197627 UUCCGAAACAUAUUUUUCUGAGUUUUCGAGAGUUUUGACCACAUUCUGGCUGCUCUUUGCAGCCUUUUCUUAACGUCUAGGUUGGCGUUUCUUCUAUGGUACUCACGCAAAAAUCCUAUG (((.((((......)))).)))((((((.((((....((((......((((((.....)))))).......((((((......)))))).......)))))))).)).))))........ ( -28.70, z-score = -2.10, R) >droSec1.super_2 2062033 120 + 7591821 GUGCGAAAUUUGUUUUUCUGAGCUUUAGAUAGUUUUGACCACAUCUUGGCUGCUCUUUGCAGCCUUUUCUUUACGUCUCCGGUGGCGUUCCUCCUUUGGAACUCUCGCAAAAAUCUUAUG .(((((..........((.(((((......))))).))((((.....((((((.....)))))).........((....)))))).(((((......)))))..)))))........... ( -33.60, z-score = -3.39, R) >droSim1.chr3L 1607011 120 + 22553184 GUGCGAAAUUUGUUUUUCUGAGCUUUAGAUAGUUUUGACCACAUCUUGGCUGCUCUUUGCAGCCUUUUCUUUACGUCUCCGGUGGCGUUCCUCCUUUGGAACUCUCGCAAAAAUCUUAUG .(((((..........((.(((((......))))).))((((.....((((((.....)))))).........((....)))))).(((((......)))))..)))))........... ( -33.60, z-score = -3.39, R) >consensus GUGCGAAAUUUGUUUUUCUGAGCUUUAGAUAGUUUUGACCACAUCUUGGCUGCUCUUUGCAGCCUUUUCUUUACGUCUACGGUGGCGUUCCUCCUUUGGAACUCACGCAAAAAUCCUAUG ................((.(((((......))))).))((.......((((((.....))))))........(((((......))))).........))..................... (-15.10 = -14.55 + -0.55)

| Location | 2,037,532 – 2,037,652 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.78 |
| Shannon entropy | 0.47252 |
| G+C content | 0.42361 |
| Mean single sequence MFE | -29.98 |
| Consensus MFE | -12.46 |
| Energy contribution | -12.72 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.42 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.725764 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
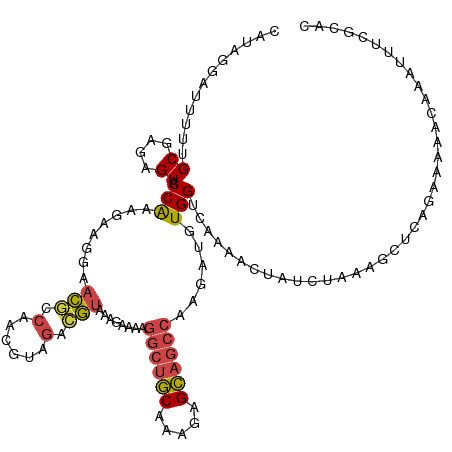
>dm3.chr3L 2037532 120 - 24543557 CAUUGGAUUUUUGCGCGAGUACCAAAGGAGGAACACCAUCGUAGACGUAAAGAAAAGGCUGCAAAGAGCAGCCAAGAUGUGGUCAAAACUAUCUAAAGCUCAGAAAAACAAAUUUCGCAC ..(((..((((((.((..((.((......)).))(((((((....)).........((((((.....)))))).....)))))..............)).))))))..)))......... ( -28.00, z-score = -1.54, R) >droAna3.scaffold_13337 22495837 120 + 23293914 CCUUGGUUUCAUGCGUGAGUUCCGAAGAAAUAAUUCAAAUAUGGGGACGCUGCUUUCACUCCAAAAAGGUACCAAAGAAUGGGUUAAACUAUCAAAGGACGAUCAAGAUCGAUACCGUAA ..((((......(((.(.(((((....................))))).)))).......))))...((((((...((.(((......)))))...)).((((....)))).)))).... ( -20.87, z-score = 1.39, R) >droEre2.scaffold_4784 2032400 120 - 25762168 CAUAGGAUUUCUGCGCGAGUACCAUAGAAGAAACGCCAGCUUAGACGUAAAGAAAAGGCUGCAAAGAGCAGCCAAGAUGUGGUCAAAACUCACGGAAGCUCAAAAAAGCAAAUUUCGAAU ....(..((((((...((((((((((......(((.(......).)))........((((((.....))))))....))))))....)))).))))))..)................... ( -28.50, z-score = -1.36, R) >droYak2.chr3L 1994235 120 - 24197627 CAUAGGAUUUUUGCGUGAGUACCAUAGAAGAAACGCCAACCUAGACGUUAAGAAAAGGCUGCAAAGAGCAGCCAGAAUGUGGUCAAAACUCUCGAAAACUCAGAAAAAUAUGUUUCGGAA .....((.((((.((.((((((((((.....((((.(......).)))).......((((((.....))))))....))))))....)))).)))))).)).((((......)))).... ( -29.30, z-score = -1.98, R) >droSec1.super_2 2062033 120 - 7591821 CAUAAGAUUUUUGCGAGAGUUCCAAAGGAGGAACGCCACCGGAGACGUAAAGAAAAGGCUGCAAAGAGCAGCCAAGAUGUGGUCAAAACUAUCUAAAGCUCAGAAAAACAAAUUUCGCAC ...........((((((((((((......)))))(((((((....)).........((((((.....)))))).....))))).............................))))))). ( -36.60, z-score = -4.32, R) >droSim1.chr3L 1607011 120 - 22553184 CAUAAGAUUUUUGCGAGAGUUCCAAAGGAGGAACGCCACCGGAGACGUAAAGAAAAGGCUGCAAAGAGCAGCCAAGAUGUGGUCAAAACUAUCUAAAGCUCAGAAAAACAAAUUUCGCAC ...........((((((((((((......)))))(((((((....)).........((((((.....)))))).....))))).............................))))))). ( -36.60, z-score = -4.32, R) >consensus CAUAGGAUUUUUGCGAGAGUACCAAAGAAGGAACGCCAACGUAGACGUAAAGAAAAGGCUGCAAAGAGCAGCCAAGAUGUGGUCAAAACUAUCUAAAGCUCAGAAAAACAAAUUUCGCAC ............((....)).(((........(((.(......).)))........((((((.....))))))......)))...................................... (-12.46 = -12.72 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:57:01 2011