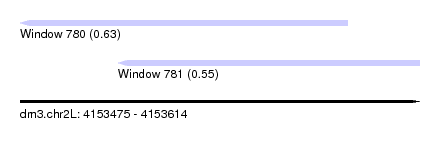
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 4,153,475 – 4,153,614 |
| Length | 139 |
| Max. P | 0.626751 |

| Location | 4,153,475 – 4,153,589 |
|---|---|
| Length | 114 |
| Sequences | 12 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 96.12 |
| Shannon entropy | 0.08578 |
| G+C content | 0.39506 |
| Mean single sequence MFE | -30.56 |
| Consensus MFE | -24.95 |
| Energy contribution | -24.81 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.28 |
| SVM RNA-class probability | 0.626751 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 4153475 114 - 23011544 UAACAUUUAAAGUCAAAUUACGGCAAAUGGUCGCAAAGUGUGUGAUUUAAAUGCACUUGGGCGACACUUUAAGACGCUCAAUUGCU-CGCUUUAUUGU----GUGGCCAUAAAAAAAGU ...........(((.......)))..(((((((((.((((.((((.......(((.(((((((.(.......).))))))).))))-)))..)))).)----))))))))......... ( -30.01, z-score = -1.86, R) >droPer1.super_1 7045939 118 + 10282868 UAACAUUUAAAGUCAAAUUACGGCAAAUGGUCGCAAAGUGUGUGAUUUAAAUGCACUUGGGCGACACUUUAAGACGCUCAAUUGCUCCGCUUUAUUGUGGGGGGGACCAUAAAAAAGG- ...........(((.......)))..((((((.(.((((((((.......))))))))(((((.(.......).))))).....((((((......))))))).))))))........- ( -33.20, z-score = -2.34, R) >dp4.chr4_group3 9925923 117 + 11692001 UAACAUUUAAAGUCAAAUUACGGCAAAUGGUCGCAAAGUGUGUGAUUUAAAUGCACUUGGGCGACACUUUAAGACGCUCAAUUGCUCCGCUUUAUUGU-GGGGGGACCAUAAAAAAGG- ...........(((.......)))..((((((((((.((((((.......))))))(((((((.(.......).)))))))))))(((((......))-)))..))))))........- ( -32.00, z-score = -2.12, R) >droAna3.scaffold_12943 725136 113 + 5039921 UAACAUUUAAAGUCAAAUUACGGCAAAUGGUCGCAAAGUGUGUGAUUUAAAUGCACUUGGGCGACACUUUAAGACGCUCAAUUGCU-CGCUUUAUUGU----GGGGCCAUAAAAAAGG- ...(((((...(((.......))))))))(((((.((((((((.......))))))))..))))).......((.((......)))-).((((.((((----(....))))).)))).- ( -29.00, z-score = -1.71, R) >droEre2.scaffold_4929 4218184 113 - 26641161 UAACAUUUAAAGUCAAAUUACGGCAAAUGGUCGCAAAGUGUGUGAUUUAAAUGCACUUGGGCGACACUUUAAGACGCUCAAUUGCU-CGCUUUAUUGU----GUGGCCAUAAAAAAGG- ...........(((.......)))..(((((((((.((((.((((.......(((.(((((((.(.......).))))))).))))-)))..)))).)----))))))))........- ( -30.01, z-score = -1.95, R) >droYak2.chr2L 4179324 113 - 22324452 UAACAUUUAAAGUCAAAUUACGGCAAAUGGUCGCAAAGUGUGUGAUUUAAAUGCACUUGGGCGACACUUUAAGACGCUCAAUUGCU-CGCUUUAUUGU----GUGGCCAUAAAAAAGG- ...........(((.......)))..(((((((((.((((.((((.......(((.(((((((.(.......).))))))).))))-)))..)))).)----))))))))........- ( -30.01, z-score = -1.95, R) >droSec1.super_5 2251818 114 - 5866729 UAACAUUUAAAGUCAAAUUACGGCAAAUGGUCGCAAAGUGUGUGAUUUAAAUGCACUUGGGCGACACUUUAAGACGCUCAAUUGCU-CGCUUUAUUGU----GUGGCCAUAAAAAAAGG ...........(((.......)))..(((((((((.((((.((((.......(((.(((((((.(.......).))))))).))))-)))..)))).)----))))))))......... ( -30.01, z-score = -1.95, R) >droSim1.chr2L 4109105 114 - 22036055 UAACAUUUAAAGUCAAAUUACGGCAAAUGGUCGCAAAGUGUGUGAUUUAAAUGCACUUGGGCGACACUUUAAGACGCUCAAUUGCU-CGCUUUAUUGU----GUGGCCAUAAAAAAAGG ...........(((.......)))..(((((((((.((((.((((.......(((.(((((((.(.......).))))))).))))-)))..)))).)----))))))))......... ( -30.01, z-score = -1.95, R) >droMoj3.scaffold_6500 2273380 114 - 32352404 UAACAUUUAAAGUCAAAUUACGGCAAAUGGUCGCAAAGUGUGUGAUUUAAAUGCACUUGGGCGACACUUUAAGACGCUCAAUUGCU-CGCUUUAUUGU----GCGGCCAUAAAAAAGCC ...........(((.......)))..(((((((((.((((.((((.......(((.(((((((.(.......).))))))).))))-)))..)))).)----))))))))......... ( -31.91, z-score = -2.03, R) >droVir3.scaffold_12963 11157942 114 + 20206255 UAACAUUUAAAGUCAAAUUACGGCAAAUGGUCGCAAAGUGUGUGAUUUAAAUGCACUUGGGCGACACUUUAAGACGCUCAAUUGCU-CGCUUUAUUGU----GCGGCCAUAAAAAAGGC ...........(((.......)))..(((((((((.((((.((((.......(((.(((((((.(.......).))))))).))))-)))..)))).)----))))))))......... ( -31.91, z-score = -2.02, R) >droGri2.scaffold_15252 12748734 108 - 17193109 UAACAUUUAAAGUCAAAUUACGGCAAAUGGUCGCAAAGUGUGUGAUUUAAAUGCACUUGGGCGACACUUUAAGACGCUCAAUUGCU-CGCUUUAUUGU----GGGGCCAUAAA------ .....................(((.....(((((.((((((((.......))))))))..)))))...................((-(((......))----)))))).....------ ( -28.70, z-score = -1.70, R) >droWil1.scaffold_180703 2164413 112 - 3946847 UAACAUUUAAAGUCAAAUUACGGCAAAUGGUCGCAAAGUGUGUGAUUUAAAUGCACUUGGGCGACACUUUAAGACGCUCAAUUGCU-CGCUUUAUUGU----GUGGCCAUAAAAAAG-- ...........(((.......)))..(((((((((.((((.((((.......(((.(((((((.(.......).))))))).))))-)))..)))).)----)))))))).......-- ( -30.01, z-score = -2.11, R) >consensus UAACAUUUAAAGUCAAAUUACGGCAAAUGGUCGCAAAGUGUGUGAUUUAAAUGCACUUGGGCGACACUUUAAGACGCUCAAUUGCU_CGCUUUAUUGU____GUGGCCAUAAAAAAGG_ ...........(((.......)))..((((((((((.((((((.......))))))(((((((.(.......).)))))))))))...((......))......))))))......... (-24.95 = -24.81 + -0.14)


| Location | 4,153,509 – 4,153,614 |
|---|---|
| Length | 105 |
| Sequences | 12 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 89.15 |
| Shannon entropy | 0.23298 |
| G+C content | 0.43031 |
| Mean single sequence MFE | -29.14 |
| Consensus MFE | -20.60 |
| Energy contribution | -20.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.81 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.548754 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 4153509 105 - 23011544 -----AUGAGGGCGAAUCAA-------CUCUUGG-CCAUAACAUUUAAAGUCAAAUUACGGCAAAUGGUCGCAAAGUGUGUGAUUUAAAUGCACUUGGGCGACACUUUAAGACGCUCA -----....(((((......-------.((((((-......(((((...(((.......))))))))(((((.((((((((.......))))))))..)))))...))))))))))). ( -28.71, z-score = -1.96, R) >droPer1.super_1 7045977 106 + 10282868 -----CUGAAGGCGAAUCAA------CCUCUUGG-CCAUAACAUUUAAAGUCAAAUUACGGCAAAUGGUCGCAAAGUGUGUGAUUUAAAUGCACUUGGGCGACACUUUAAGACGCUCA -----.....((((......------((....))-........((((((((.......((.....))(((((.((((((((.......))))))))..))))))))))))).)))).. ( -26.00, z-score = -1.02, R) >dp4.chr4_group3 9925960 106 + 11692001 -----CUGAAGGCGAAUCAA------CCUCUUGG-CCAUAACAUUUAAAGUCAAAUUACGGCAAAUGGUCGCAAAGUGUGUGAUUUAAAUGCACUUGGGCGACACUUUAAGACGCUCA -----.....((((......------((....))-........((((((((.......((.....))(((((.((((((((.......))))))))..))))))))))))).)))).. ( -26.00, z-score = -1.02, R) >droAna3.scaffold_12943 725169 105 + 5039921 -----CUGAAGGCGAAUCAA-------CUCUUGG-CCAUAACAUUUAAAGUCAAAUUACGGCAAAUGGUCGCAAAGUGUGUGAUUUAAAUGCACUUGGGCGACACUUUAAGACGCUCA -----.....(((.((....-------...)).)-))......((((((((.......((.....))(((((.((((((((.......))))))))..)))))))))))))....... ( -25.50, z-score = -1.03, R) >droEre2.scaffold_4929 4218217 105 - 26641161 -----AUGAGGGCGAAUCAA-------CUCUUGG-CCAUAACAUUUAAAGUCAAAUUACGGCAAAUGGUCGCAAAGUGUGUGAUUUAAAUGCACUUGGGCGACACUUUAAGACGCUCA -----....(((((......-------.((((((-......(((((...(((.......))))))))(((((.((((((((.......))))))))..)))))...))))))))))). ( -28.71, z-score = -1.96, R) >droYak2.chr2L 4179357 105 - 22324452 -----AUGAGGGCGAAUCAG-------CUCUUGG-CCAUAACAUUUAAAGUCAAAUUACGGCAAAUGGUCGCAAAGUGUGUGAUUUAAAUGCACUUGGGCGACACUUUAAGACGCUCA -----....(((((.....(-------(.....)-).......((((((((.......((.....))(((((.((((((((.......))))))))..))))))))))))).))))). ( -29.50, z-score = -1.77, R) >droSec1.super_5 2251852 105 - 5866729 -----AUGAGGGCGAAUCAA-------CUCUUGG-CCAUAACAUUUAAAGUCAAAUUACGGCAAAUGGUCGCAAAGUGUGUGAUUUAAAUGCACUUGGGCGACACUUUAAGACGCUCA -----....(((((......-------.((((((-......(((((...(((.......))))))))(((((.((((((((.......))))))))..)))))...))))))))))). ( -28.71, z-score = -1.96, R) >droSim1.chr2L 4109139 105 - 22036055 -----AUGAGGGCGAAUCAA-------CUCUUGG-CCAUAACAUUUAAAGUCAAAUUACGGCAAAUGGUCGCAAAGUGUGUGAUUUAAAUGCACUUGGGCGACACUUUAAGACGCUCA -----....(((((......-------.((((((-......(((((...(((.......))))))))(((((.((((((((.......))))))))..)))))...))))))))))). ( -28.71, z-score = -1.96, R) >droMoj3.scaffold_6500 2273414 101 - 32352404 ----------UUUGAGACAU------CUCGGGGG-UCAUAACAUUUAAAGUCAAAUUACGGCAAAUGGUCGCAAAGUGUGUGAUUUAAAUGCACUUGGGCGACACUUUAAGACGCUCA ----------...(((....------)))(((.(-((....(((((...(((.......))))))))(((((.((((((((.......))))))))..))))).......))).))). ( -31.90, z-score = -3.90, R) >droVir3.scaffold_12963 11157976 112 + 20206255 -----UUGACGUCGCGACGUCCGAGGCCCGGGGG-UCAUAACAUUUAAAGUCAAAUUACGGCAAAUGGUCGCAAAGUGUGUGAUUUAAAUGCACUUGGGCGACACUUUAAGACGCUCA -----.(((((((..(((.((((.....)))).)-)).......(((((((.......((.....))(((((.((((((((.......))))))))..)))))))))))))))).))) ( -37.50, z-score = -2.32, R) >droGri2.scaffold_15252 12748762 118 - 17193109 UUAGAGAGAAGGAGAGGGGAGAGGUGGUGGGGAGCCCAUAACAUUUAAAGUCAAAUUACGGCAAAUGGUCGCAAAGUGUGUGAUUUAAAUGCACUUGGGCGACACUUUAAGACGCUCA .............(((.(.((((((.(((((...))))).)).......(((.......))).....(((((.((((((((.......))))))))..))))).))))....).))). ( -32.50, z-score = -2.22, R) >droWil1.scaffold_180703 2164445 108 - 3946847 -----AGGAGAAUGAAUCGAC----GACCCUGGG-CCAUAACAUUUAAAGUCAAAUUACGGCAAAUGGUCGCAAAGUGUGUGAUUUAAAUGCACUUGGGCGACACUUUAAGACGCUCA -----.(..((.....))..)----......(((-(.....(((((...(((.......))))))))(((((.((((((((.......))))))))..)))))..........)))). ( -25.90, z-score = -0.60, R) >consensus _____AUGAGGGCGAAUCAA_______CUCUUGG_CCAUAACAUUUAAAGUCAAAUUACGGCAAAUGGUCGCAAAGUGUGUGAUUUAAAUGCACUUGGGCGACACUUUAAGACGCUCA ...........................................((((((((.......((.....))(((((.((((((((.......))))))))..)))))))))))))....... (-20.60 = -20.60 + -0.00)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:15:31 2011