| Sequence ID | dm3.chr3L |
|---|---|
| Location | 2,026,268 – 2,026,362 |
| Length | 94 |
| Max. P | 0.500000 |

| Location | 2,026,268 – 2,026,362 |
|---|---|
| Length | 94 |
| Sequences | 8 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 80.63 |
| Shannon entropy | 0.37229 |
| G+C content | 0.47081 |
| Mean single sequence MFE | -25.84 |
| Consensus MFE | -15.11 |
| Energy contribution | -16.92 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.58 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>dm3.chr3L 2026268 94 + 24543557 -GAGGGUAGUGAGUAAGCAGCGAUUGAGACCUGAACAAAUUAAUCGCCAUUCGGGGCUGAACUGAAAG-------UCAGGUUUGACCGAGUUUACACAAUCU -..((((..((.((((((.((((((((............))))))))...((((..(....(((....-------.)))....).)))))))))).)))))) ( -23.80, z-score = -0.83, R) >droSim1.chr3L 1595833 94 + 22553184 -GAGGGAAGUGAGUAAGCAGCGAUUGAGACCUGAACAAAUUAAUCGCCAUUCGGGGCUAAACUGAAAG-------UCAGGUUUGACCGAGUUUACACACUCU -...(((.(((.((((((.((((((((............))))))))...((((...((((((.....-------...)))))).)))))))))).)))))) ( -27.90, z-score = -2.25, R) >droSec1.super_2 2050978 94 + 7591821 -GAGGAAAGUGGGUAAGCAGCGAUUGAGACCUGAACAAAUUAAUCGCCAUUCGGGGUUGAACUGAAAG-------UCAGGUUUGACCGAGUUUACACACUCU -......((((.((((((.((((((((............)))))))).......(((..((((.....-------...))))..)))..)))))).)))).. ( -29.10, z-score = -2.40, R) >droYak2.chr3L 1982163 94 + 24197627 -GAGGGAAGUGAGUAAGCAGCGAUUGAGACCUGAACAAAUUAAUCGCCAUUCGGUGCUGAACUGAAAG-------UCAGGUUUGACCGAGUUUACACACUCU -...(((.(((.((((((.((((((((............))))))))...(((((.(....(((....-------.)))....)))))))))))).)))))) ( -28.90, z-score = -2.41, R) >droEre2.scaffold_4784 2020930 94 + 25762168 -GAGGGAAGUGAGCAAGCAGCGAUUGAGACCUGAACAAAUUAAUCGCCAUUCGGUGCUGAACUGAAAG-------UCAGGUUUGACCGAGUUUACACACUCU -(((....((((((.....((((((((............))))))))...(((((.(....(((....-------.)))....))))))))))))...))). ( -26.50, z-score = -1.49, R) >droAna3.scaffold_13337 22484162 101 - 23293914 -GCGAGGCGGGGGCAAGCAGCGAUUGAGACUUGAACAAAUUAAUCGCCAUUCGGGGCAGGGCCGACAGAGCAUAGUCAGGUUUGACCGAGUUUACACACUCU -..((((.(..(((.....((((((((............))))))))...((((....((((((((........))).)))))..)))))))..).).))). ( -28.00, z-score = -0.62, R) >dp4.chrXR_group6 11924534 80 + 13314419 CGUGGGAAA-GCGUAAGCAGCGAUUGAGACUUGAACAAAUUAAUCGCC----GGGACU-----------------CUGGUUUUGACCGAGUUUACAGAGUUU .........-((....)).((((((((............)))))))).----.(((((-----------------(((..(((....)))....)))))))) ( -20.00, z-score = -0.77, R) >droPer1.super_20 1845560 78 + 1872136 CGUGGGAAA-GCGUAAGCAGCGAUUGAGACUUGAACAAAUUAAUCGCC----GGGACU-----------------CUGGUUUUGACCGAGUUUACACUUU-- .(((.....-((....)).((((((((............)))))))).----.(((((-----------------(.(((....))))))))).)))...-- ( -22.50, z-score = -1.93, R) >consensus _GAGGGAAGUGAGUAAGCAGCGAUUGAGACCUGAACAAAUUAAUCGCCAUUCGGGGCUGAACUGAAAG_______UCAGGUUUGACCGAGUUUACACACUCU ............((((((.((((((((............))))))))...((((...((((((...............)))))).))))))))))....... (-15.11 = -16.92 + 1.81)

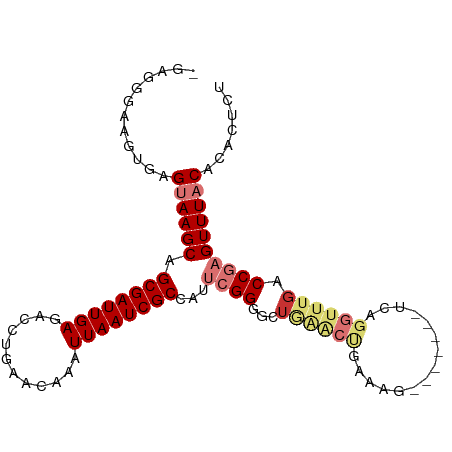
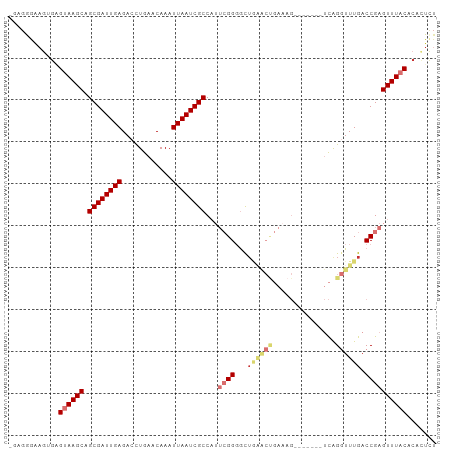
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:56:57 2011