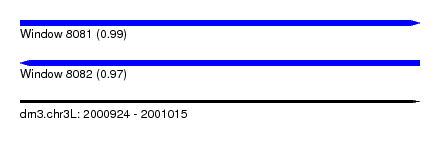
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 2,000,924 – 2,001,015 |
| Length | 91 |
| Max. P | 0.992080 |

| Location | 2,000,924 – 2,001,015 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 90.27 |
| Shannon entropy | 0.16469 |
| G+C content | 0.48393 |
| Mean single sequence MFE | -22.64 |
| Consensus MFE | -19.64 |
| Energy contribution | -20.12 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.92 |
| Structure conservation index | 0.87 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.992080 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
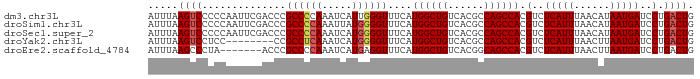
>dm3.chr3L 2000924 91 + 24543557 AUUUAAGUCCCCCAAUUCGACCCGCCCCAAAUCAUUGGGUUUCAUGGCUGUCACGCCAGCCACGUCUCAUUUAACAUAAUGAUCCUGACUG .....((((.........(((....(((((....))))).....((((((......)))))).)))(((((......)))))....)))). ( -22.20, z-score = -3.10, R) >droSim1.chr3L 1570306 91 + 22553184 AUUUAAGUCCCCCAAUUCGACCCGCCCCAAAUUAUGGGGUUUCAUGGCUGUCACGCCAGCCACGUCUCAUUUAACAUAAUGAUCCUGACUG .....((((.........(((..((((((.....))))))....((((((......)))))).)))(((((......)))))....)))). ( -26.10, z-score = -3.79, R) >droSec1.super_2 2025990 91 + 7591821 AUUUAAGUCCCCCAAUUCGACCCGCCCCAAAUCAUGGGGUUUCAUGGCUGUCACGCCAGCCACGUCUCAUUUAACAUAAUGAUCCUGACUG .....((((.........(((..((((((.....))))))....((((((......)))))).)))(((((......)))))....)))). ( -26.10, z-score = -3.69, R) >droYak2.chr3L 1955971 83 + 24197627 AUUUAAGUCCUCC--------CCGCCUCAAAUCAUGGGGUUUCAUGGCUGUCACGCCAGCCACGUCUCAUUUAACUUAAUGAUCCUGACUG .....((((....--------..((((((.....))))))....((((((......)))))).(..(((((......)))))..).)))). ( -20.80, z-score = -2.52, R) >droEre2.scaffold_4784 1995743 84 + 25762168 AUUUAAGCCCUA-------ACCCGCCCCAAAUCAUGAGGUUUCAUGGCUGUCACGGCAGCCACGUCUCAUUUAACUUAAUGAUCCUGACUG ............-------....(((.((.....)).)))....(((((((....))))))).((((((((......)))))....))).. ( -18.00, z-score = -1.52, R) >consensus AUUUAAGUCCCCCAAUUCGACCCGCCCCAAAUCAUGGGGUUUCAUGGCUGUCACGCCAGCCACGUCUCAUUUAACAUAAUGAUCCUGACUG .....((((..............((((((.....))))))....((((((......)))))).(..(((((......)))))..).)))). (-19.64 = -20.12 + 0.48)
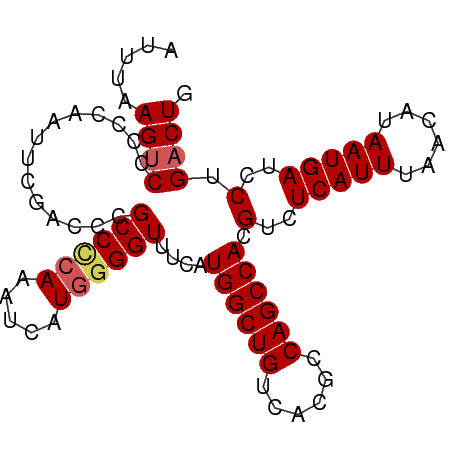
| Location | 2,000,924 – 2,001,015 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 90.27 |
| Shannon entropy | 0.16469 |
| G+C content | 0.48393 |
| Mean single sequence MFE | -28.88 |
| Consensus MFE | -24.80 |
| Energy contribution | -24.92 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.972546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr3L 2000924 91 - 24543557 CAGUCAGGAUCAUUAUGUUAAAUGAGACGUGGCUGGCGUGACAGCCAUGAAACCCAAUGAUUUGGGGCGGGUCGAAUUGGGGGACUUAAAU .((((....(((((......)))))..((((((((......))))))))...((((((((((((...))))))..)))))).))))..... ( -29.60, z-score = -2.55, R) >droSim1.chr3L 1570306 91 - 22553184 CAGUCAGGAUCAUUAUGUUAAAUGAGACGUGGCUGGCGUGACAGCCAUGAAACCCCAUAAUUUGGGGCGGGUCGAAUUGGGGGACUUAAAU .((((....(((((......)))))..((((((((......))))))))...(((((..((((((......)))))))))))))))..... ( -31.00, z-score = -2.67, R) >droSec1.super_2 2025990 91 - 7591821 CAGUCAGGAUCAUUAUGUUAAAUGAGACGUGGCUGGCGUGACAGCCAUGAAACCCCAUGAUUUGGGGCGGGUCGAAUUGGGGGACUUAAAU .((((....(((((......)))))..((((((((......))))))))...((((((((((((...)))))))...)))))))))..... ( -31.40, z-score = -2.55, R) >droYak2.chr3L 1955971 83 - 24197627 CAGUCAGGAUCAUUAAGUUAAAUGAGACGUGGCUGGCGUGACAGCCAUGAAACCCCAUGAUUUGAGGCGG--------GGAGGACUUAAAU .((((....(((((......)))))..((((((((......))))))))...((((............))--------))..))))..... ( -26.20, z-score = -2.55, R) >droEre2.scaffold_4784 1995743 84 - 25762168 CAGUCAGGAUCAUUAAGUUAAAUGAGACGUGGCUGCCGUGACAGCCAUGAAACCUCAUGAUUUGGGGCGGGU-------UAGGGCUUAAAU .((((..((((.(((((((..(((((.((((((((......))))))))....))))))))))))....)))-------)..))))..... ( -26.20, z-score = -1.99, R) >consensus CAGUCAGGAUCAUUAUGUUAAAUGAGACGUGGCUGGCGUGACAGCCAUGAAACCCCAUGAUUUGGGGCGGGUCGAAUUGGGGGACUUAAAU .((((....(((((......)))))..((((((((......))))))))...(((((.....)))))...............))))..... (-24.80 = -24.92 + 0.12)

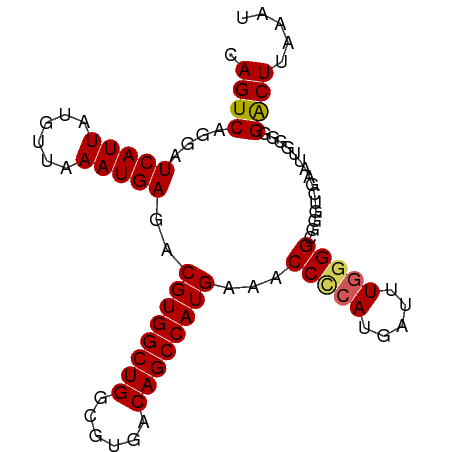
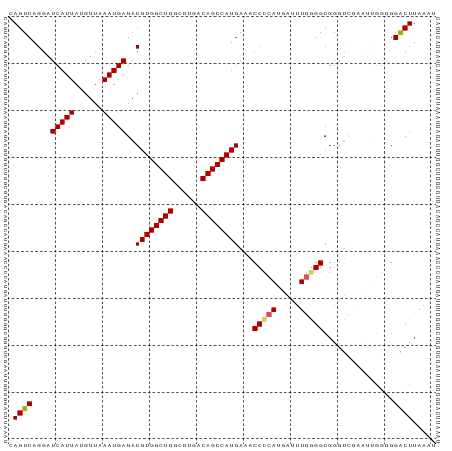
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:56:54 2011