
| Sequence ID | dm3.chr3L |
|---|---|
| Location | 1,984,991 – 1,985,099 |
| Length | 108 |
| Max. P | 0.940853 |

| Location | 1,984,991 – 1,985,099 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 134 |
| Reading direction | forward |
| Mean pairwise identity | 71.87 |
| Shannon entropy | 0.42706 |
| G+C content | 0.41377 |
| Mean single sequence MFE | -27.44 |
| Consensus MFE | -20.10 |
| Energy contribution | -20.46 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.47 |
| SVM RNA-class probability | 0.940853 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
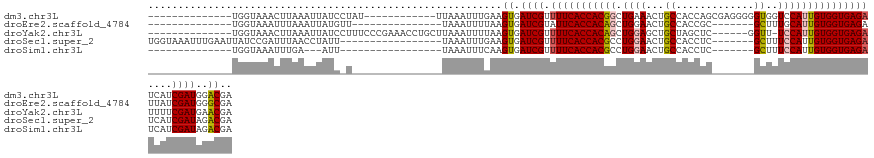
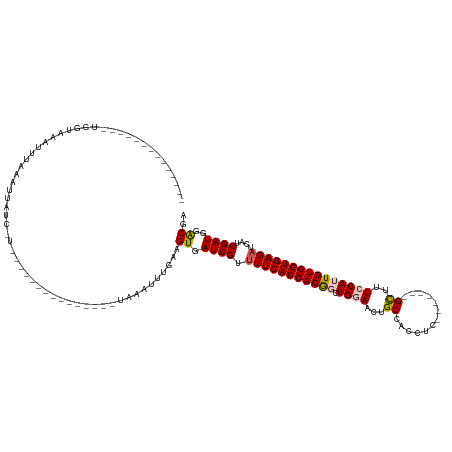
>dm3.chr3L 1984991 108 + 24543557 --------------UGGUAAACUUAAAUUAUCCUAU------------UUAAAUUUGAAGUGAUCGUUUUCACCACGGCUGAAACUGCCACCAGCGAGGGGGUGGUCCAUUGUGGUGAGAUCAUCGAUGGACGA --------------...((((.((((((......))------------)))).))))..((.((((.(((((((((((.((..((..((.((......))))..)).)))))))))))))....)))).).).. ( -29.80, z-score = -0.71, R) >droEre2.scaffold_4784 1974046 98 + 25762168 --------------UGGUAAAUUUAAAUUAUGUU---------------UAAAUUUUAAGUGAUCGUAUUCACCACAGCUGGAACUGCCACCGC-------GCUUUGCAUUGUGGUGAGAUUAUCGAUGGGCGA --------------..((((((((((((...)))---------------)))))))....(.((((..((((((((((.((.((..((......-------)).)).)))))))))))).....)))).))).. ( -22.10, z-score = -0.08, R) >droYak2.chr3L 1939707 113 + 24197627 --------------UGGUAAACUUAAAUUAUCCUUUCCCGAAACCUGCUUAAAUUUUAAGUGAUCGUUUUCACCACAGCUGGAGCUGCUAGCUC------GGUU-UCCAUUGUGGUGAGAUUUUCGAUGAACGA --------------.(((((.......)))))......((......(((((.....))))).((((.(((((((((((.((((((((......)------)).)-)))))))))))))))....))))...)). ( -29.10, z-score = -1.84, R) >droSec1.super_2 2010179 110 + 7591821 UGGUAAAUUUGAAUUAUCCGAUUUAACCUAUU-----------------UAAAUUUGAAGUGAUCGUUUUCACCACGCCUGGAACUGCCACCUC-------GCUUUCCAUUGUGGUGAGAUCAUCGAUAGACGA .((((((((.((....)).))))).)))....-----------------..........((.((((.((((((((((..(((((..((......-------)).))))).))))))))))....))))..)).. ( -28.90, z-score = -2.57, R) >droSim1.chr3L 1554150 93 + 22553184 --------------UGGUAAAUUUGA---AUU-----------------UAAAUUUCAAGUGAUCGUUUUCACCACGCCUGGAACUGCCACCUC-------GCUUUCCAUUGUGGUGAGAUCAUCGAUAGACGA --------------......((((((---(..-----------------.....))))))).((((.((((((((((..(((((..((......-------)).))))).))))))))))....))))...... ( -27.30, z-score = -2.73, R) >consensus ______________UGGUAAAUUUAAAUUAUC_U_______________UAAAUUUGAAGUGAUCGUUUUCACCACGGCUGGAACUGCCACCUC_______GCUUUCCAUUGUGGUGAGAUCAUCGAUGGACGA ...........................................................((.((((.(((((((((((.((((...((.............))..)))))))))))))))....))))..)).. (-20.10 = -20.46 + 0.36)
| Location | 1,984,991 – 1,985,099 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 134 |
| Reading direction | reverse |
| Mean pairwise identity | 71.87 |
| Shannon entropy | 0.42706 |
| G+C content | 0.41377 |
| Mean single sequence MFE | -25.86 |
| Consensus MFE | -13.75 |
| Energy contribution | -14.83 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.728613 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
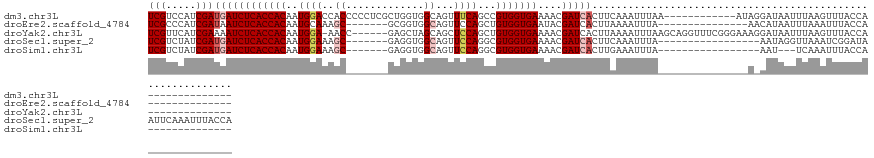
>dm3.chr3L 1984991 108 - 24543557 UCGUCCAUCGAUGAUCUCACCACAAUGGACCACCCCCUCGCUGGUGGCAGUUUCAGCCGUGGUGAAAACGAUCACUUCAAAUUUAA------------AUAGGAUAAUUUAAGUUUACCA-------------- (((.....)))((((((((((((..(((((((((........)))))....))))...)))))))....)))))....((((((((------------((......))))))))))....-------------- ( -28.80, z-score = -2.10, R) >droEre2.scaffold_4784 1974046 98 - 25762168 UCGCCCAUCGAUAAUCUCACCACAAUGCAAAGC-------GCGGUGGCAGUUCCAGCUGUGGUGAAUACGAUCACUUAAAAUUUA---------------AACAUAAUUUAAAUUUACCA-------------- .........(((....((((((((..((.....-------))(.(((.....))).))))))))).....))).....(((((((---------------((.....)))))))))....-------------- ( -21.10, z-score = -1.69, R) >droYak2.chr3L 1939707 113 - 24197627 UCGUUCAUCGAAAAUCUCACCACAAUGGA-AACC------GAGCUAGCAGCUCCAGCUGUGGUGAAAACGAUCACUUAAAAUUUAAGCAGGUUUCGGGAAAGGAUAAUUUAAGUUUACCA-------------- (((.....)))...((((..(.....)((-((((------..(((.(((((....)))))(((((......))))).........))).))))))))))..((.(((.......))))).-------------- ( -25.80, z-score = -1.06, R) >droSec1.super_2 2010179 110 - 7591821 UCGUCUAUCGAUGAUCUCACCACAAUGGAAAGC-------GAGGUGGCAGUUCCAGGCGUGGUGAAAACGAUCACUUCAAAUUUA-----------------AAUAGGUUAAAUCGGAUAAUUCAAAUUUACCA (((.....)))((((((((((((..(((((.((-------......))..)))))...)))))))....)))))...........-----------------....(((.((((..((....))..))))))). ( -28.40, z-score = -2.05, R) >droSim1.chr3L 1554150 93 - 22553184 UCGUCUAUCGAUGAUCUCACCACAAUGGAAAGC-------GAGGUGGCAGUUCCAGGCGUGGUGAAAACGAUCACUUGAAAUUUA-----------------AAU---UCAAAUUUACCA-------------- .......((((((((((((((((..(((((.((-------......))..)))))...)))))))....))))).))))......-----------------...---............-------------- ( -25.20, z-score = -1.90, R) >consensus UCGUCCAUCGAUGAUCUCACCACAAUGGAAAGC_______GAGGUGGCAGUUCCAGCCGUGGUGAAAACGAUCACUUAAAAUUUA_______________A_AAUAAUUUAAAUUUACCA______________ (((.....)))((((((((((((..(((((....................)))))...)))))))....)))))............................................................ (-13.75 = -14.83 + 1.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 22:56:53 2011